null
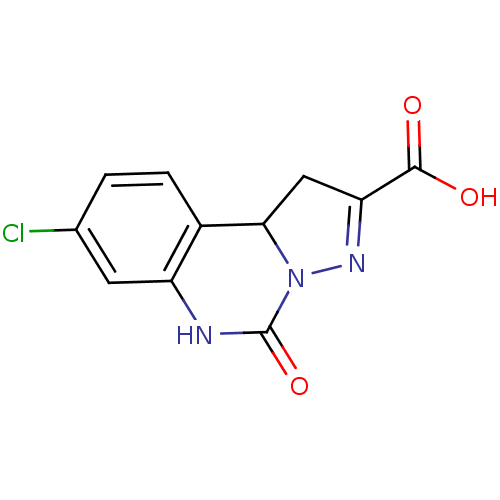
SMILES OC(=O)C1=NN2C(C1)c1ccc(Cl)cc1NC2=O
InChI Key InChIKey=OWXURGXAWBPHEV-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50110213
Found 2 hits for monomerid = 50110213
Affinity DataKi: 240nMAssay Description:Binding selectivity towards N-methyl-D-aspartate glutamate receptor 1 was determined by using [3H]- glycine/NMDAMore data for this Ligand-Target Pair
Affinity DataKi: 240nMAssay Description:Displacement of [3H]-Glycine from N-methyl-D-aspartate glutamate receptor 1 in rat cortical membranesMore data for this Ligand-Target Pair