null
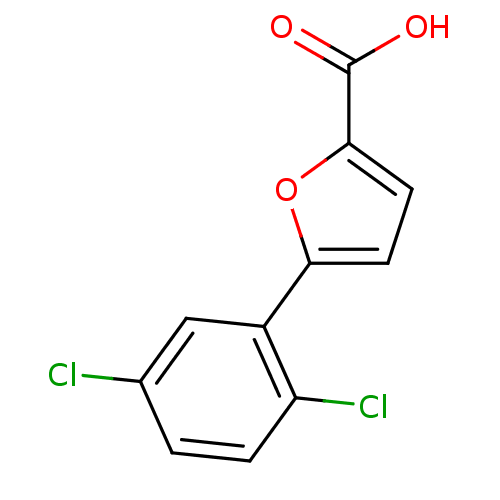
SMILES OC(=O)c1ccc(o1)-c1cc(Cl)ccc1Cl
InChI Key InChIKey=ATAZLMGGQQLRBC-UHFFFAOYSA-N
PDB links: 1 PDB ID matches this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50175451
Found 4 hits for monomerid = 50175451
TargetMethionine aminopeptidase(Escherichia coli (strain K12))
Graduate School of Chinese Academy of Sciences
Curated by ChEMBL
Graduate School of Chinese Academy of Sciences
Curated by ChEMBL
Affinity DataIC50: 1.26E+5nMAssay Description:Inhibitory activity against Co(II) form of Escherichia coli MetAPMore data for this Ligand-Target Pair
In DepthDetails
Article
BindingDB Entry DOI: 10.7270/Q2HH6JNVPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)
BindingDB Entry DOI: 10.7270/Q2HH6JNVPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)TargetMethionine aminopeptidase(Escherichia coli (strain K12))
Graduate School of Chinese Academy of Sciences
Curated by ChEMBL
Graduate School of Chinese Academy of Sciences
Curated by ChEMBL
Affinity DataIC50: 8.40E+4nMAssay Description:Inhibitory activity against Fe(II) form of Escherichia coli MetAPMore data for this Ligand-Target Pair
In DepthDetails
Article
BindingDB Entry DOI: 10.7270/Q2HH6JNVPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)
BindingDB Entry DOI: 10.7270/Q2HH6JNVPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)TargetMethionine aminopeptidase(Escherichia coli (strain K12))
Graduate School of Chinese Academy of Sciences
Curated by ChEMBL
Graduate School of Chinese Academy of Sciences
Curated by ChEMBL
Affinity DataIC50: 693nMAssay Description:Inhibitory activity against Mn(II) form of Escherichia coli MetAPMore data for this Ligand-Target Pair
In DepthDetails
Article
BindingDB Entry DOI: 10.7270/Q2HH6JNVPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)
BindingDB Entry DOI: 10.7270/Q2HH6JNVPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)TargetMethionine aminopeptidase(Escherichia coli (strain K12))
Graduate School of Chinese Academy of Sciences
Curated by ChEMBL
Graduate School of Chinese Academy of Sciences
Curated by ChEMBL
Affinity DataIC50: 1.73E+5nMAssay Description:Inhibitory activity against Ni(II) form of Escherichia coli MetAPMore data for this Ligand-Target Pair
In DepthDetails
Article
BindingDB Entry DOI: 10.7270/Q2HH6JNVPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)
BindingDB Entry DOI: 10.7270/Q2HH6JNVPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)