null
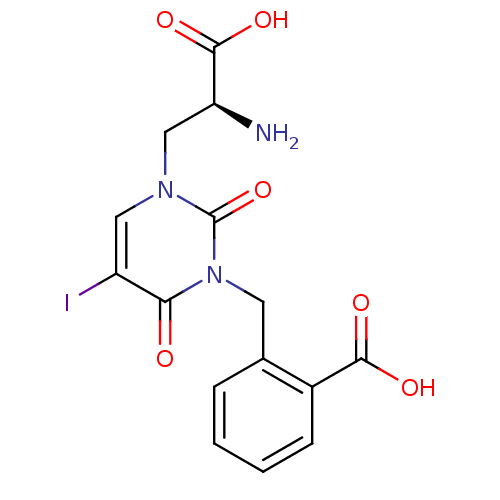
SMILES N[C@@H](Cn1cc(I)c(=O)n(Cc2ccccc2C(O)=O)c1=O)C(O)=O
InChI Key InChIKey=HZEJBGVIAXRMKT-NSHDSACASA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50178145
Found 3 hits for monomerid = 50178145
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]kainate from rat GLUK6 receptor expressed in HEK293 cellsMore data for this Ligand-Target Pair
Affinity DataKd: 209nMAssay Description:Activity at native GLUK5 kainate receptor assessed as antagonism of kainite-induced depolarization of neonatal anaesthetised rat dorsal root fibresMore data for this Ligand-Target Pair
Affinity DataKd: 209nMAssay Description:Antagonism on GLUK5 containing kainate induced deploarization of isolated neonatal rat dorsal root C-fibersMore data for this Ligand-Target Pair