null
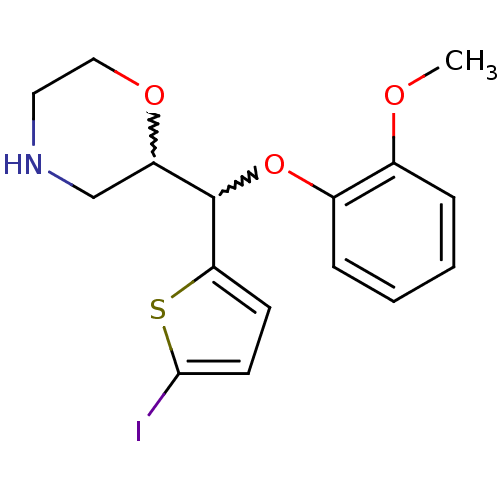
SMILES COc1ccccc1OC(C1CNCCO1)c1ccc(I)s1
InChI Key InChIKey=TUMPGFWCYJFORL-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50198228
Found 3 hits for monomerid = 50198228
Affinity DataKi: 79.6nMAssay Description:Displacement of [3H]nisoxetine from NET in rat brain membranesMore data for this Ligand-Target Pair
TargetSodium-dependent serotonin transporter(Rattus norvegicus (rat))
Institute for Degenerative Diseases
Curated by ChEMBL
Institute for Degenerative Diseases
Curated by ChEMBL
Affinity DataKi: 141nMAssay Description:Displacement of [3H]paroxetine from SERT in brain membranesMore data for this Ligand-Target Pair
TargetSodium-dependent dopamine transporter(Rattus norvegicus (rat))
Institute for Degenerative Diseases
Curated by ChEMBL
Institute for Degenerative Diseases
Curated by ChEMBL
Affinity DataKi: 210nMAssay Description:Displacement of [3H]GBR from DAT in rat brain membranesMore data for this Ligand-Target Pair