null
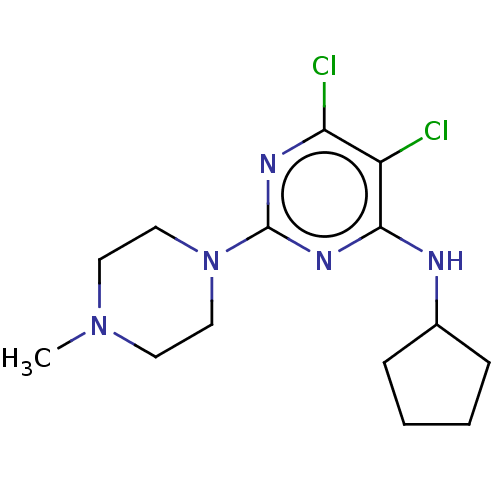
SMILES CN1CCN(CC1)c1nc(Cl)c(Cl)c(NC2CCCC2)n1
InChI Key InChIKey=JJUHBOAGMJQKNV-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50226322
Found 3 hits for monomerid = 50226322
TargetAlpha-2A adrenergic receptor [16-465]/Alpha-2B adrenergic receptor/Alpha-2C adrenergic receptor(RAT)
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataKi: 97nMAssay Description:In vitro binding affinity was measured as the inhibition of [3H]idazoxan to alpha-2 adrenergic receptors of rat cortical membranesMore data for this Ligand-Target Pair
TargetAlpha-2A adrenergic receptor [16-465]/Alpha-2B adrenergic receptor/Alpha-2C adrenergic receptor(RAT)
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataKi: 120nMAssay Description:In vitro binding affinity was measured as the inhibition of [3H]clonidine binding to alpha-2 adrenergic receptor of rat cortical membranesMore data for this Ligand-Target Pair
Affinity DataKi: 240nMAssay Description:In vitro binding affinity was measured as the inhibition of [3H]WB-4101 binding to alpha-1 adrenergic receptor of rat cortical membranesMore data for this Ligand-Target Pair