null
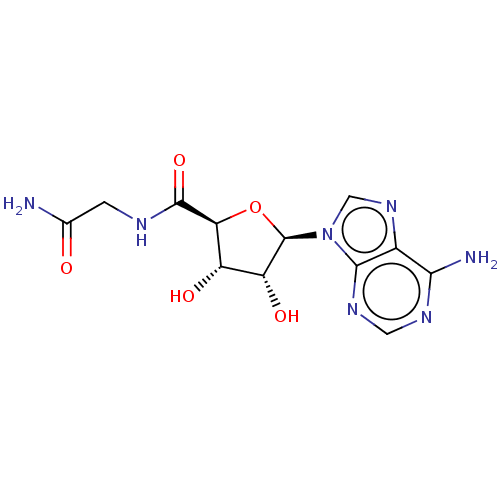
SMILES NC(=O)CNC(=O)[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c(N)ncnc12
InChI Key
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50552894
Found 4 hits for monomerid = 50552894
Affinity DataKi: 1.90E+3nMAssay Description:Displacement of FITC-geldanamycin from HSP90alpha (unknown origin) after 24 hrs by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.00E+4nMAssay Description:Displacement of FITC-geldanamycin from GRP94 (unknown origin) after 24 hrs by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKd: 3.20E+4nMAssay Description:Displacement of FITC-geldanamycin from HSP90alpha (unknown origin) after 24 hrs by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKd: >5.00E+4nMAssay Description:Displacement of FITC-geldanamycin from GRP94 (unknown origin) after 24 hrs by fluorescence polarization assayMore data for this Ligand-Target Pair