null
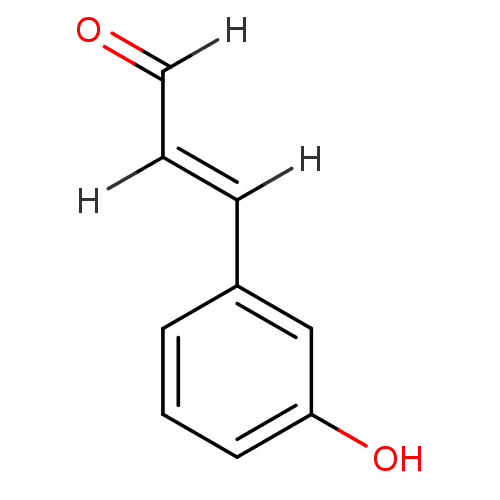
SMILES Oc1cccc(\C=C\C=O)c1
InChI Key InChIKey=DCHPWNOJPJRSEA-DUXPYHPUSA-N
PDB links: 33 PDB IDs contain this monomer as substructures. 33 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 7569
Found 2 hits for monomerid = 7569
TargetCyclin-dependent kinase 4/G1/S-specific cyclin-D1(Homo sapiens (Human))
Korea Research Institute of Bioscience and Biotechnology
Korea Research Institute of Bioscience and Biotechnology
Affinity DataIC50: 4.50E+4nMpH: 7.2 T: 2°CAssay Description:Kinase activities were assayed in buffers containing substrate, enzyme, and inhibitor at 30 °C in the presence of 100 uM ATP/ [gamma-33P] ATP. 3...More data for this Ligand-Target Pair
TargetCyclin-dependent kinase 1/G2/mitotic-specific cyclin-B(Marthasterias glacialis (starfish))
Korea Research Institute of Bioscience and Biotechnology
Korea Research Institute of Bioscience and Biotechnology
Affinity DataIC50: 3.00E+4nMpH: 7.2 T: 2°CAssay Description:Kinase activities were assayed in buffers containing substrate, enzyme, and inhibitor at 30 °C in the presence of 15 uM ATP/ [gamma-32P] ATP. 32...More data for this Ligand-Target Pair