null
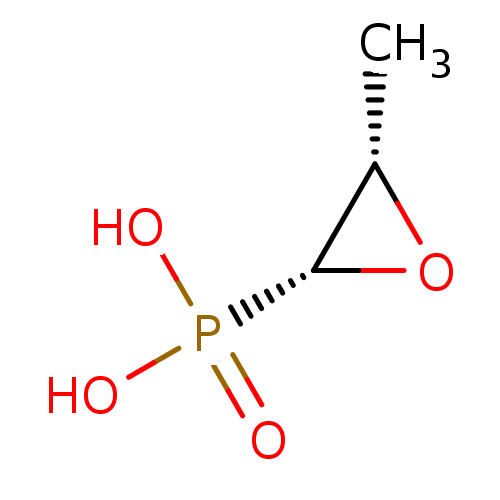
SMILES C[C@@H]1O[C@@H]1P(O)(O)=O
InChI Key InChIKey=YMDXZJFXQJVXBF-STHAYSLISA-N
PDB links: 14 PDB IDs match this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 6 hits for monomerid = 50024894
Found 6 hits for monomerid = 50024894
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 118nMAssay Description:Inhibition of Escherichia coli K12 Mur A in presence of UNAGMore data for this Ligand-Target Pair
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 100nMAssay Description:Inhibition of Escherichia coli MurA expressed in Escherichia coli BL21(lamdaDE3) using UNAG and PEP as substrate incubated for 10 mins prior to PEP a...More data for this Ligand-Target Pair
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 8.80E+3nMAssay Description:Inhibition of Escherichia coli K-12 MBP-fused C-terminal His-tagged MurA (6508 to 7768 residues) using UNAG as substrate preincubated for 10 mins fol...More data for this Ligand-Target Pair
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Pseudomonas aeruginosa (G-proteobacteria))
Saarland University
Curated by ChEMBL
Saarland University
Curated by ChEMBL
Affinity DataIC50: 1.05E+4nMAssay Description:Inhibition of Pseudomonas aeruginosa PAO1293 MurA in presence of UNAGMore data for this Ligand-Target Pair