null
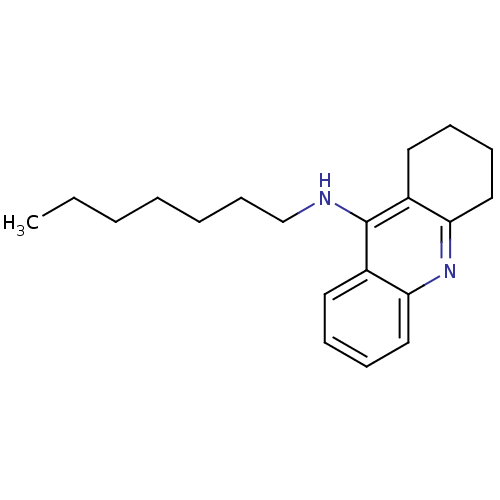
SMILES CCCCCCCNc1c2CCCCc2nc2ccccc12
InChI Key InChIKey=HQRYMINYXXVYNF-UHFFFAOYSA-N
PDB links: 5 PDB IDs contain this monomer as substructures. 5 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 10442
Found 2 hits for monomerid = 10442
TargetCarboxylic ester hydrolase(Rattus norvegicus (rat))
Hong Kong University of Science and Technology
Hong Kong University of Science and Technology
Affinity DataIC50: 340nMAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at...More data for this Ligand-Target Pair
Affinity DataIC50: 454nMpH: 7.4 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at...More data for this Ligand-Target Pair