null
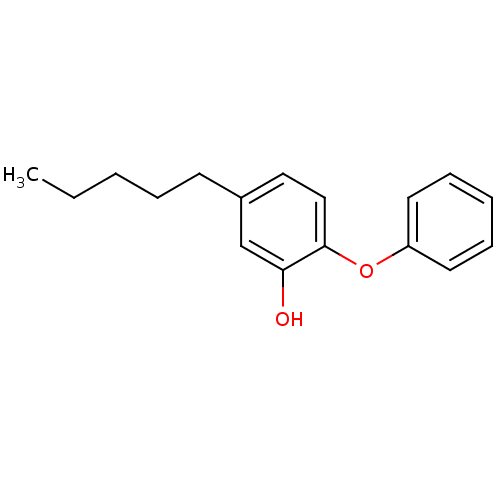
SMILES CCCCCc1ccc(Oc2ccccc2)c(O)c1
InChI Key InChIKey=OJLYTHOKCYLPMA-UHFFFAOYSA-N
PDB links: 2 PDB IDs match this monomer. 1 PDB ID contains this monomer as substructures. 1 PDB ID contains inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 6 hits for monomerid = 16296
Found 6 hits for monomerid = 16296
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0400nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
Affinity DataKi: 1.30nM ΔG°: -12.1kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition constant binding to E-NAD+More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
SUNY Stony Brook
SUNY Stony Brook
Affinity DataKi: 11.8nM ΔG°: -10.7kcal/molepH: 6.8 T: 2°CAssay Description:Inhibition constants (Ki) were calculated by determining the kcat and Km (DDCoA) values at several fixed inhibitor concentrations. The inhibition dat...More data for this Ligand-Target Pair
In DepthDetails
Article
BindingDB Entry DOI: 10.7270/Q2K35RWKPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)
BindingDB Entry DOI: 10.7270/Q2K35RWKPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)TargetEnoyl-[acyl-carrier-protein] reductase [NADH] [T276S](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataIC50: 1.00E+5nMAssay Description:IC50 values for the inhibition of wildtype and mutant forms of ypFabV were performed by adding varying concentrations of inhibitor dissolved in dimet...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
SUNY Stony Brook
SUNY Stony Brook
Affinity DataIC50: 17nMpH: 6.8 T: 2°CAssay Description:The assay measured the NADH-dependent catalysis of a DD-CoA substrate as a decrease in 340 nm absorbance resulting from conversion of NADH to NAD. Th...More data for this Ligand-Target Pair
In DepthDetails
Article
BindingDB Entry DOI: 10.7270/Q2K35RWKPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)
BindingDB Entry DOI: 10.7270/Q2K35RWKPubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataIC50: 1.00E+3nMAssay Description:IC50 values for the inhibition of wildtype and mutant forms of ypFabV were performed by adding varying concentrations of inhibitor dissolved in dimet...More data for this Ligand-Target Pair