null
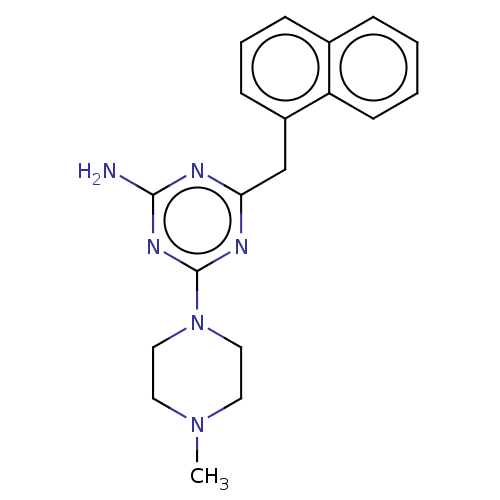
SMILES CN1CCN(CC1)c1nc(N)nc(Cc2cccc3ccccc23)n1
InChI Key InChIKey=ZJCMJVPWMYOZEM-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 189352
Found 3 hits for monomerid = 189352
Target5-hydroxytryptamine receptor 6(Homo sapiens (Human))
Department of Technology and Biotechnology of Drugs, Jagiellonian University, Medical College, Medyczna 9, PL 30-688 Kraków, Poland. Electronic address: dlazewska@cm-uj.krakow.pl.
Curated by ChEMBL
Department of Technology and Biotechnology of Drugs, Jagiellonian University, Medical College, Medyczna 9, PL 30-688 Kraków, Poland. Electronic address: dlazewska@cm-uj.krakow.pl.
Curated by ChEMBL
Affinity DataKi: 20nMAssay Description:Displacement of [3H]-LSD from human 5-HT6R expressed in human HeLa cellsMore data for this Ligand-Target Pair
Affinity DataKi: 5.60E+3nMAssay Description:Displacement of [3H]histamine from human H4R expressed in Sf9 cell membranes co-expressed with G protein Gai2 and Gb1gamma2 incubated for 60 mins by ...More data for this Ligand-Target Pair
Affinity DataKi: 5.60E+3nM ΔG°: -6.66kcal/molepH: 7.4 T: 2°CAssay Description:Prior to the experiments, cell membranes were sedimented by a 10 min centrifugation at 4 °C
and 16 0009x g and resuspended in binding buffer (12...More data for this Ligand-Target Pair