null
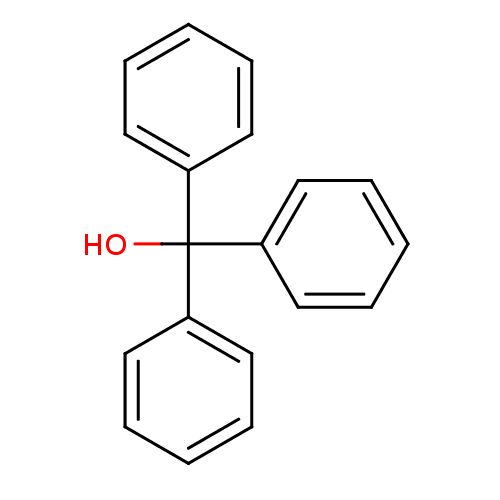
SMILES OC(c1ccccc1)(c1ccccc1)c1ccccc1
InChI Key InChIKey=LZTRCELOJRDYMQ-UHFFFAOYSA-N
PDB links: 6 PDB IDs contain this monomer as substructures. 6 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 23784
Found 3 hits for monomerid = 23784
TargetDeoxyuridine 5'-triphosphate nucleotidohydrolase(Plasmodium falciparum)
Cardiff University
Curated by ChEMBL
Cardiff University
Curated by ChEMBL
Affinity DataKi: >1.00E+6nMAssay Description:Inhibition of Plasmodium falciparum dUTPaseMore data for this Ligand-Target Pair
TargetDeoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial(Homo sapiens (Human))
Cardiff University
Curated by ChEMBL
Cardiff University
Curated by ChEMBL
Affinity DataKi: >1.00E+6nMAssay Description:Inhibition of human recombinant dUTPaseMore data for this Ligand-Target Pair
Affinity DatapH: 6.9 T: 2°CAssay Description:The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E...More data for this Ligand-Target Pair