null
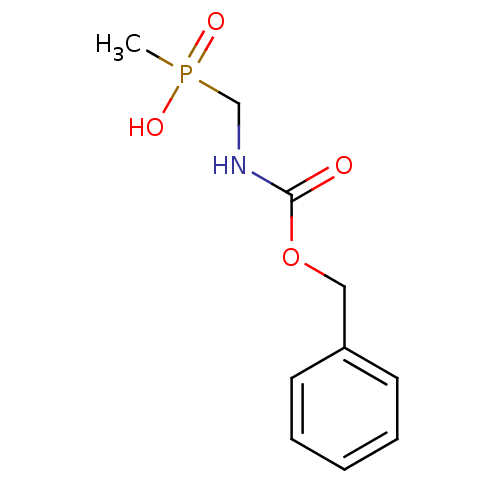
SMILES CP(O)(=O)CNC(=O)OCc1ccccc1
InChI Key InChIKey=WJYZLHFBKNWWDZ-UHFFFAOYSA-N
PDB links: 3 PDB IDs contain this monomer as substructures. 3 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 24968
Found 2 hits for monomerid = 24968
Affinity DataKi: 4.30E+4nM ΔG°: -6.05kcal/mole IC50: 1.23E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 6.50E+4nM ΔG°: -5.80kcal/mole IC50: 3.19E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair