null
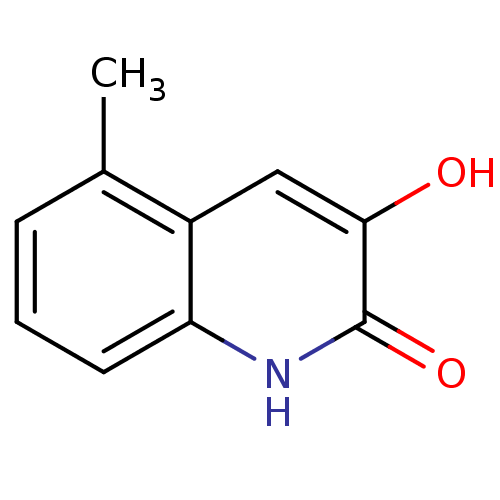
SMILES Cc1cccc2[nH]c(=O)c(O)cc12
InChI Key InChIKey=NRNHMRFPCJTZAW-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 31160
Found 3 hits for monomerid = 31160
Affinity DataIC50: 16nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 8.47E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 5.46E+4nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair