null
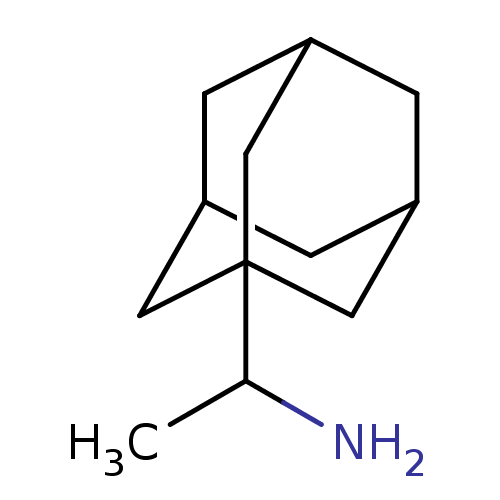
SMILES CC(N)C12CC3CC(CC(C3)C1)C2
InChI Key InChIKey=UBCHPRBFMUDMNC-UHFFFAOYSA-N
PDB links: 3 PDB IDs match this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 11 hits for monomerid = 50216627
Found 11 hits for monomerid = 50216627
TargetSolute carrier family 22 member 2(Homo sapiens (Human))
University of California
Curated by ChEMBL
University of California
Curated by ChEMBL
Affinity DataIC50: 4.40E+3nMAssay Description:Inhibition of human OCT2-mediated ASP+ uptake expressed in HEK293 cells after 3 mins by fluorescence assayMore data for this Ligand-Target Pair
TargetMultidrug and toxin extrusion protein 2(Homo sapiens (Human))
University of California
Curated by ChEMBL
University of California
Curated by ChEMBL
Affinity DataIC50: 2.88E+5nMAssay Description:Inhibition of human MATE2K-mediated ASP+ uptake expressed in HEK293 cells after 1.5 mins by fluorescence assayMore data for this Ligand-Target Pair
TargetMultidrug and toxin extrusion protein 1(Homo sapiens (Human))
University of California
Curated by ChEMBL
University of California
Curated by ChEMBL
Affinity DataIC50: 7.30E+3nMAssay Description:Inhibition of human MATE1-mediated ASP+ uptake expressed in HEK293 cells after 1.5 mins by fluorescence assayMore data for this Ligand-Target Pair
TargetMatrix protein 2(Influenza A virus (A/Udorn/307/1972(H3N2)))
National and Kapodistrian University of Athens
Curated by ChEMBL
National and Kapodistrian University of Athens
Curated by ChEMBL
Affinity DataKd: >1.00E+7nMAssay Description:Binding affinity to Influenza A virus A/Udorn/72 M2 proton channel S31N mutant expressed in Xenopus laevis oocytes at pH 5.5 by two-electrode voltage...More data for this Ligand-Target Pair
TargetMatrix protein 2(Influenza A virus (A/Udorn/307/1972(H3N2)))
National and Kapodistrian University of Athens
Curated by ChEMBL
National and Kapodistrian University of Athens
Curated by ChEMBL
Affinity DataKd: 7.00E+3nMAssay Description:Binding affinity to Influenza A virus A/Udorn/72 wild-type M2 proton channel expressed in Xenopus laevis oocytes at pH 5.5 by two-electrode voltage c...More data for this Ligand-Target Pair
In DepthDetails
TargetMatrix protein 2(Influenza A virus (A/Udorn/307/1972(H3N2)))
National and Kapodistrian University of Athens
Curated by ChEMBL
National and Kapodistrian University of Athens
Curated by ChEMBL
Affinity DataKd: 510nMAssay Description:Binding affinity to Influenza A virus A/Udorn/72 wild-type M2 proton channel by isothermal calorimetric titrationMore data for this Ligand-Target Pair
Affinity DataKd: 16nMAssay Description:Binding affinity to Influenza A Weybridge(H7N7) virus Matrix protein 2 by spectrophotometryMore data for this Ligand-Target Pair
TargetPolymerase acidic protein(Hepatitis C virus)
Saint Petersburg State University
Curated by ChEMBL
Saint Petersburg State University
Curated by ChEMBL
Affinity DataIC50: 1.10E+4nMAssay Description:Inhibition of Influenza A virus (A/Puerto Rico/8/34(H1N1)) PA endonuclease infected in MDCK cells assessed as reduction in virus titer treated with c...More data for this Ligand-Target Pair
Affinity DataIC50: 30nMAssay Description:The cytopathic effect (CPE) inhibition test is used for measuring the antiviral effect. Monolayer cells in 96-well plates are inoculated with 0.1 ml ...More data for this Ligand-Target Pair
TargetMatrix protein 2(Influenza A virus (A/Udorn/307/1972(H3N2)))
National and Kapodistrian University of Athens
Curated by ChEMBL
National and Kapodistrian University of Athens
Curated by ChEMBL
Affinity DataIC50: 1.08E+4nMAssay Description:Inhibition of Influenza A virus A/Udorn/72 wild-type M2 proton channel expressed in Xenopus laevis oocytes assessed as blockade of inward currents at...More data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 3 hits for monomerid = 50216627
Found 3 hits for monomerid = 50216627
ITC DataΔG°: -6.30kcal/mole −TΔS°: 0.521kcal/mole ΔH°: -6.83kcal/mole logk: 3.52E+4
pH: 7.0 T: 30.10°C
pH: 7.0 T: 30.10°C
Host BDBM36127(6-Amino-6-deoxy-beta-cyclodextrin (am-beta-CD) | 6...)
BDBM36127(6-Amino-6-deoxy-beta-cyclodextrin (am-beta-CD) | 6...)
Universidade de Santiago de Compostela
 BDBM36127(6-Amino-6-deoxy-beta-cyclodextrin (am-beta-CD) | 6...)
BDBM36127(6-Amino-6-deoxy-beta-cyclodextrin (am-beta-CD) | 6...)Universidade de Santiago de Compostela
ITC DataΔG°: -5.94kcal/mole −TΔS°: 0.0362kcal/mole ΔH°: -5.98kcal/mole logk: 1.93E+4
pH: 7.0 T: 30.10°C
pH: 7.0 T: 30.10°C
ITC DataΔG°: -3.30kcal/mole −TΔS°: 0.847kcal/mole ΔH°: -4.13kcal/mole logk: 241
pH: 7.0 T: 30.10°C
pH: 7.0 T: 30.10°C