null
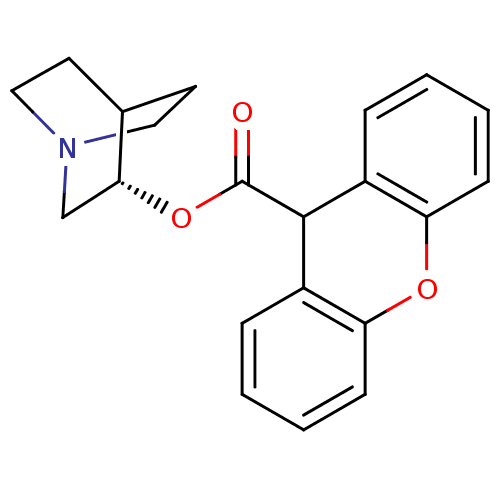
SMILES O=C(O[C@@H]1CN2CCC1CC2)C1c2ccccc2Oc2ccccc12
InChI Key InChIKey=DAIWBMZPSVQRCH-LJQANCHMSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50367741
Found 4 hits for monomerid = 50367741
Affinity DataKi: 5.20nMAssay Description:Binding affinity against Muscarinic acetylcholine receptor M1 by displacement of [3H]pirenzepine in bovine striatumMore data for this Ligand-Target Pair
Affinity DataKi: 27nMAssay Description:Evaluated for the phosphatidyl inositol turnover at Muscarinic acetylcholine receptor M1 in rat cortexMore data for this Ligand-Target Pair
Affinity DataKi: 74nMAssay Description:Binding affinity against Muscarinic acetylcholine receptor M2 by displacement of [3H]QNB in rat myocardiumMore data for this Ligand-Target Pair
Affinity DataKi: 1.66E+3nMAssay Description:Evaluated for the inhibition of adenylate cyclase at M2 receptor in rat heartMore data for this Ligand-Target Pair