null
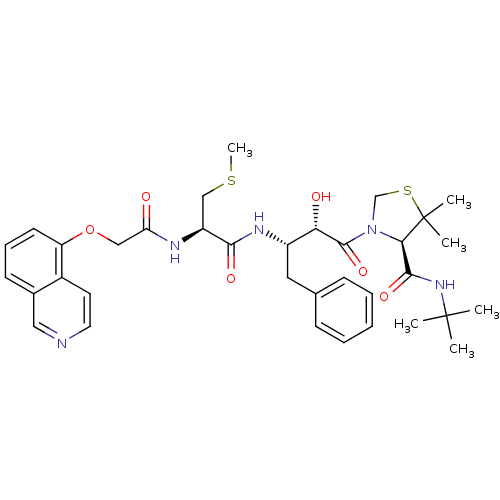
SMILES CSC[C@H](NC(=O)COc1cccc2cnccc12)C(=O)N[C@@H](Cc1ccccc1)[C@H](O)C(=O)N1CSC(C)(C)[C@H]1C(=O)NC(C)(C)C
InChI Key InChIKey=AHWCYDXQIXZVRK-RPQLRNILSA-N
PDB links: 1 PDB ID contains inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 719
Found 3 hits for monomerid = 719
TargetDimer of Gag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
National Cancer Institute
National Cancer Institute
Affinity DataKi: 0.00230nM ΔG°: -16.5kcal/molepH: 6.2 T: 2°CAssay Description:Inhibition constants were determined by a fluorometric assay with the fluorogenic substrate Arg-Glu(EDANS)-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Lys(DABCYL)-Ar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
National Cancer Institute
National Cancer Institute
Affinity DataKi: 0.0880nM ΔG°: -14.3kcal/molepH: 6.0 T: 2°CAssay Description:Sensitivity of HIV-1 protease activity to protease inhibitors was determined by a peptide substrate cleavage assay. Substrates and cleavage fragments...More data for this Ligand-Target Pair
Affinity DataKi: 36nMAssay Description:Inhibition of Plasmodium falciparum plasmepsin 2More data for this Ligand-Target Pair