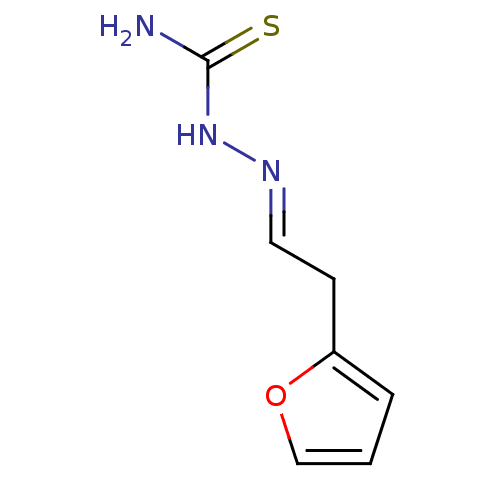
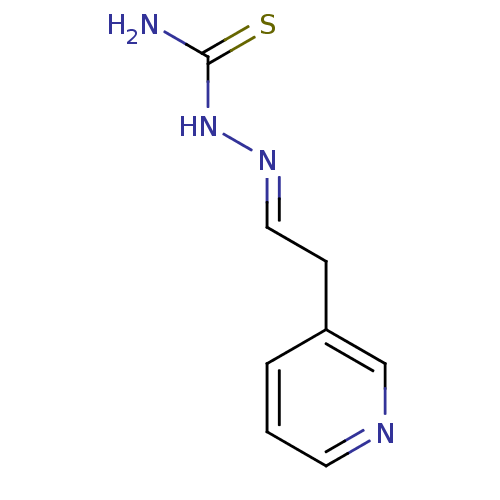
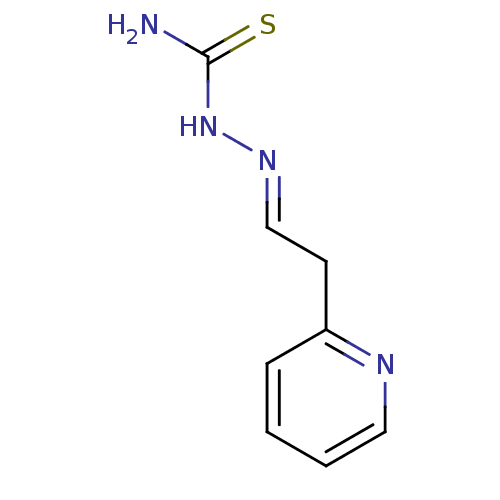
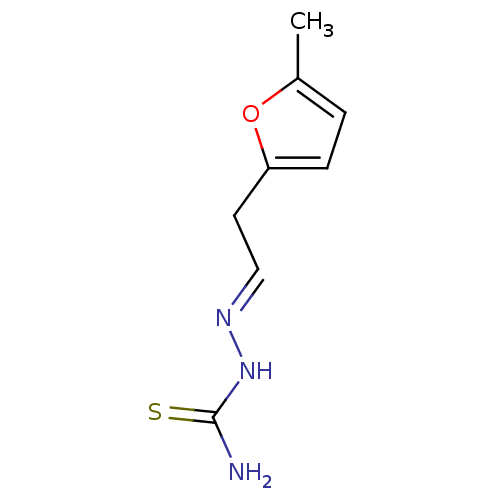
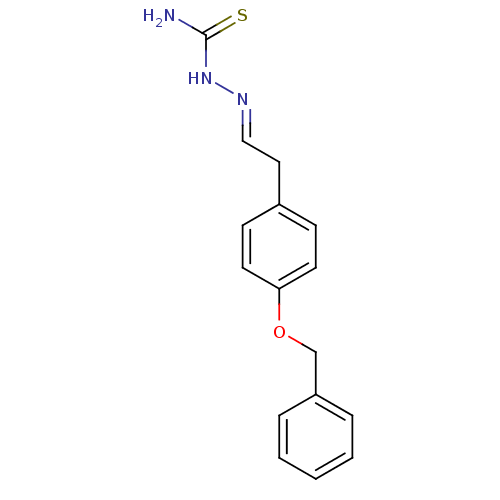
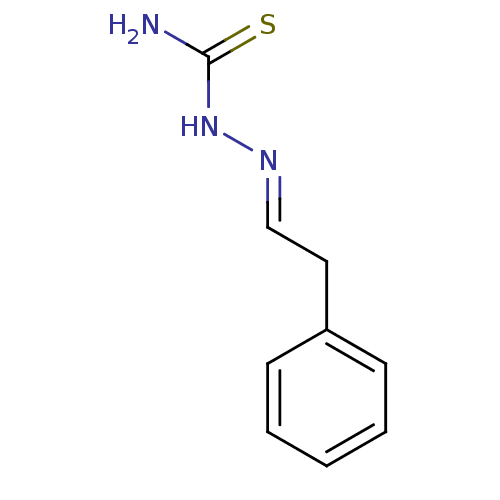
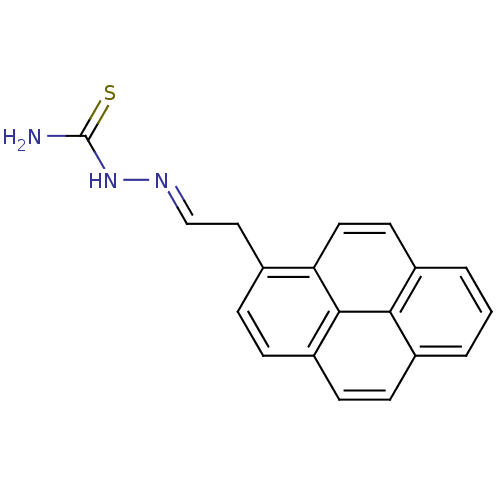
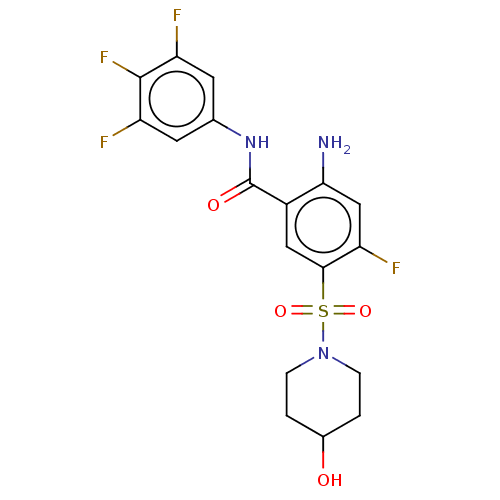
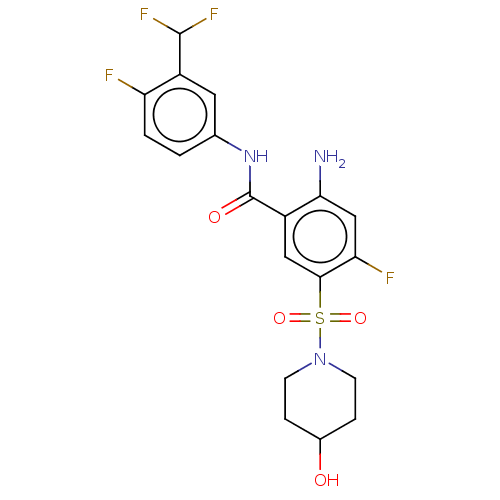
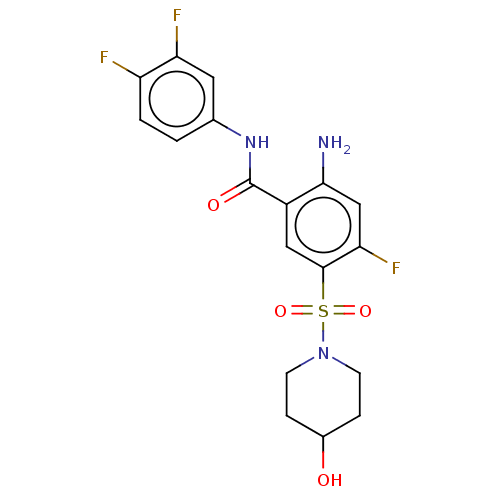
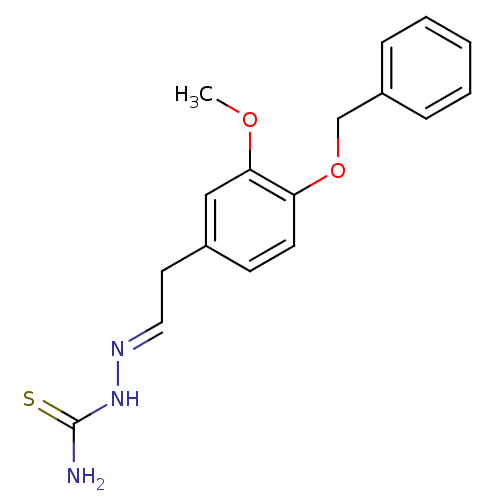
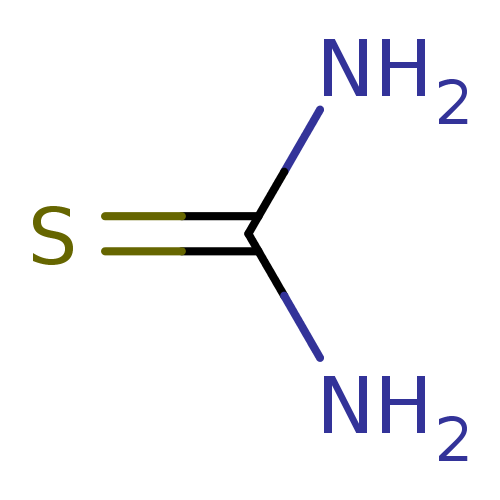Affinity DataIC50: 580nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.41E+3nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.65E+3nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
Affinity DataIC50: 3.23E+3nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
Affinity DataIC50: 3.64E+3nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
Affinity DataIC50: 4.24E+3nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
Affinity DataIC50: 4.84E+3nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
TargetPotassium voltage-gated channel subfamily H member 2(Homo sapiens (Human))
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Affinity DataIC50: 8.78E+3nMAssay Description:Inhibition of human ERG expressed in human HEK293 cells by patch clamp assayMore data for this Ligand-Target Pair
TargetPotassium voltage-gated channel subfamily H member 2(Homo sapiens (Human))
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Affinity DataIC50: 1.00E+4nMAssay Description:Inhibition of human ERG by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetPotassium voltage-gated channel subfamily H member 2(Homo sapiens (Human))
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Affinity DataIC50: 1.09E+4nMAssay Description:Inhibition of human ERG by competitive fluorescence polarization assayMore data for this Ligand-Target Pair
TargetCytochrome P450 2C9(Homo sapiens (Human))
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Affinity DataIC50: 1.70E+4nMAssay Description:Inhibition of human CYP2C9 using tolubutamide as substrate by LC/MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1.83E+4nMAssay Description:Inhibition of human liver microsome CYP2C19 using S-mephenytoin as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1.90E+4nMAssay Description:Inhibition of human liver microsome CYP2C19 using S-mephenytoin as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.08E+4nMAssay Description:Inhibition of human liver microsome CYP3A4 using sorafenib as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+4nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.31E+4nMAssay Description:Inhibition of human CYP2C19 using S-mephenytoin as substrate by LC/MS/MS analysisMore data for this Ligand-Target Pair
TargetCytochrome P450 2C9(Homo sapiens (Human))
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of human liver microsome CYP2C9 using tolbutamide as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
TargetCytochrome P450 2C9(Homo sapiens (Human))
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Affinity DataIC50: 2.52E+4nMAssay Description:Inhibition of human liver microsome CYP2C9 using tolbutamide as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.67E+4nMAssay Description:Inhibition of human liver microsome CYP2D6 using dextromethorphan as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.78E+4nMAssay Description:Inhibition of human CYP3A4 using sorafenib as substrate by LC/MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.93E+4nMAssay Description:Inhibition of human liver microsome CYP2D6 using dextromethorphan as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 3.26E+4nMAssay Description:Inhibition of human CYP2D6 using dextromethorphan as substrate by LC/MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 4.10E+4nMAssay Description:Inhibition of human liver microsome CYP3A4 using sorafenib as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 4.55E+4nMAssay Description:Inhibition of human liver microsome CYP1A2 using phenacetin as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair
TargetPotassium voltage-gated channel subfamily H member 2(Homo sapiens (Human))
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Korea Research Institute of Chemical Technology
Curated by ChEMBL
Affinity DataIC50: 5.00E+4nMAssay Description:Inhibition of human ERG by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataIC50: 7.51E+4nMpH: 8.2Assay Description:Urease inhibition activity was determined by indophenol method. In brief, each 140 無 assay reaction contained 40 無 buffer (100 mM urea, 0.01 mM K2H...More data for this Ligand-Target Pair
Affinity DataIC50: 8.11E+4nMAssay Description:Inhibition of human CYP1A2 using phenacetin as substrate by LC/MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of human liver microsome CYP1A2 using phenacetin as substrate incubated for 20 mins by LC-MS/MS with HPLC analysisMore data for this Ligand-Target Pair