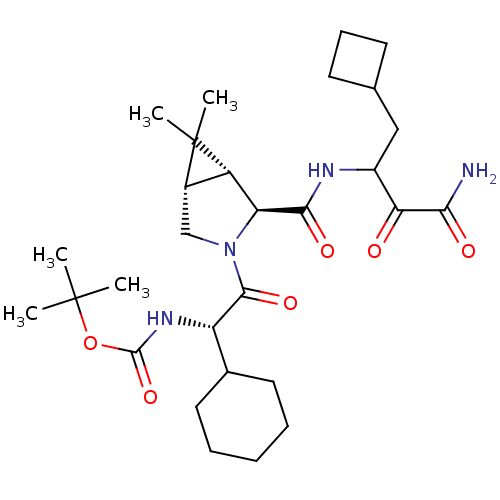
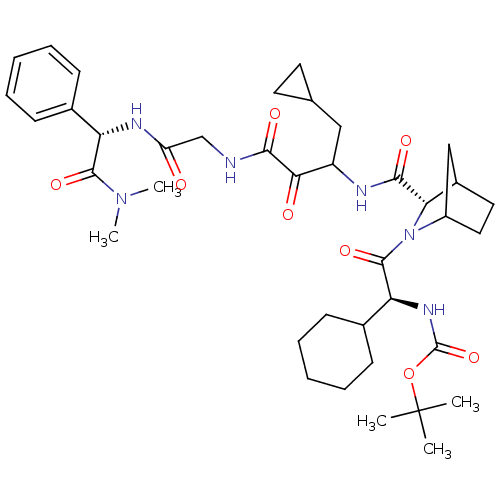
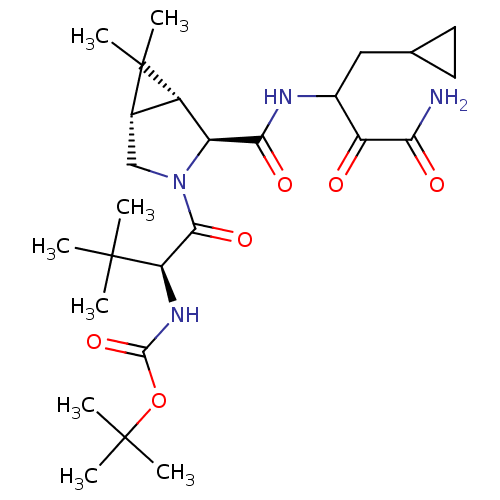
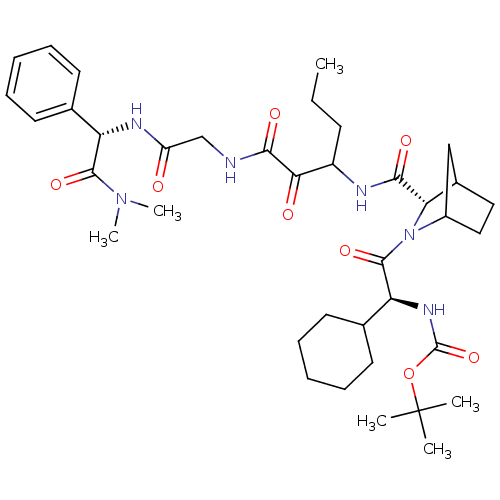
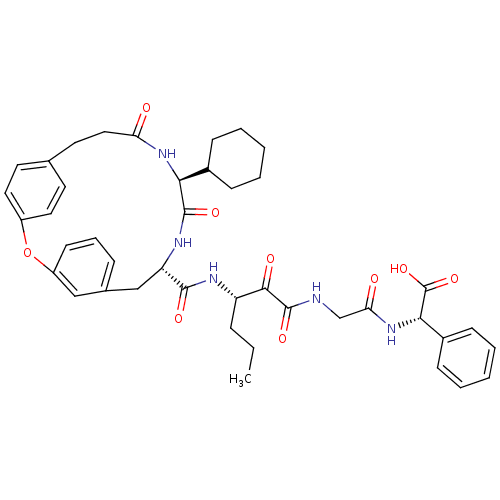
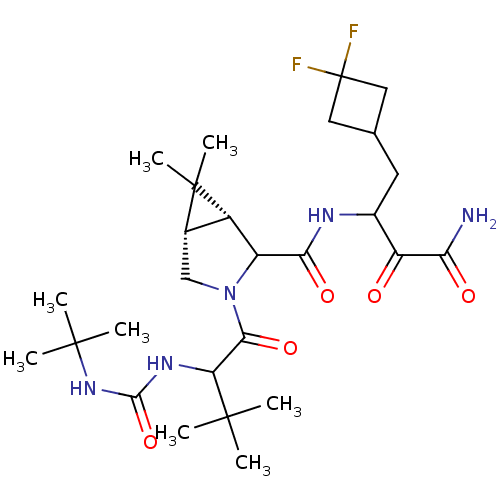
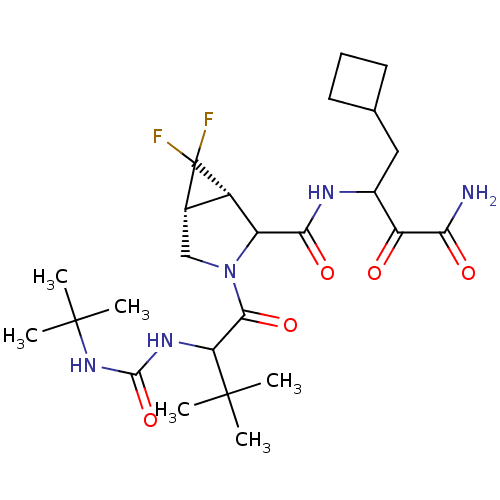
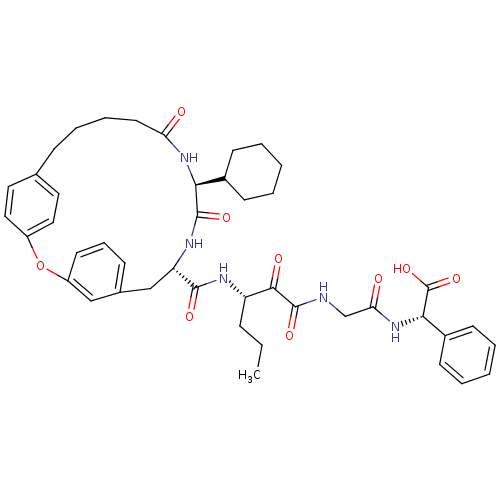
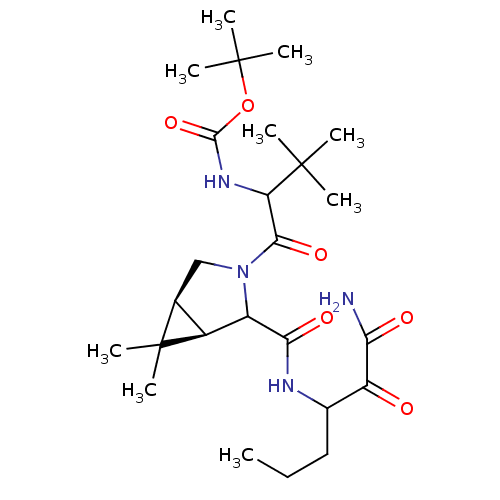
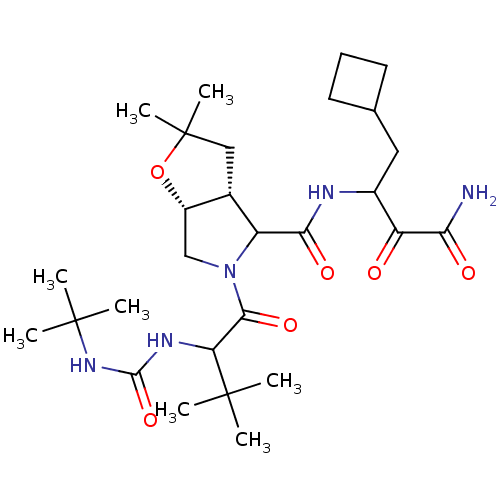
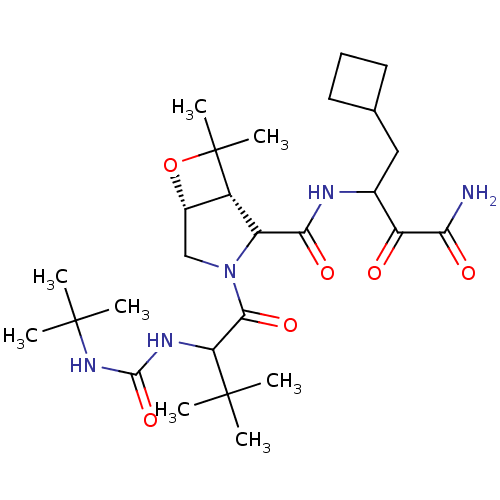
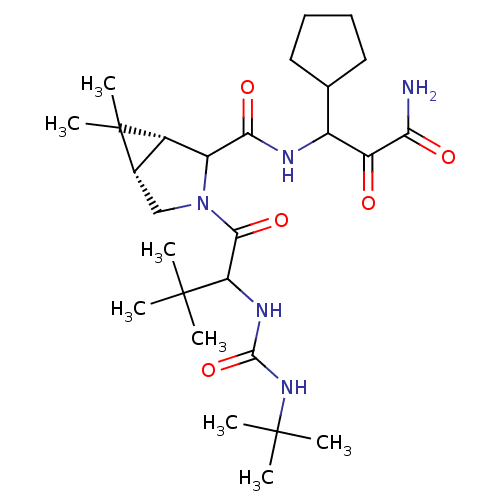
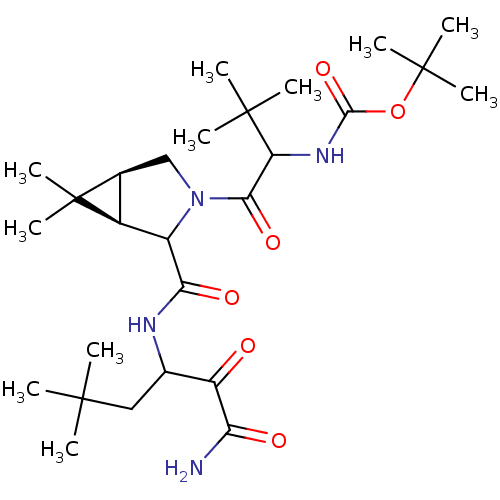
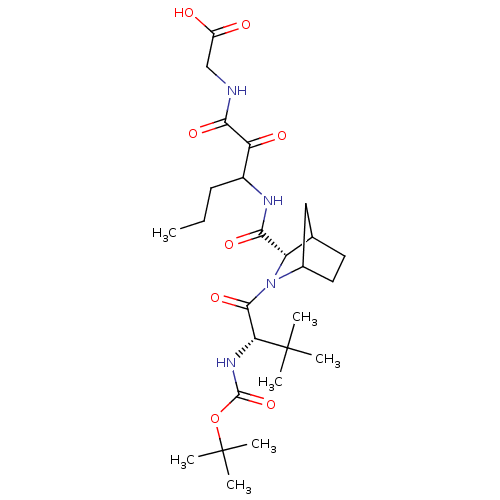
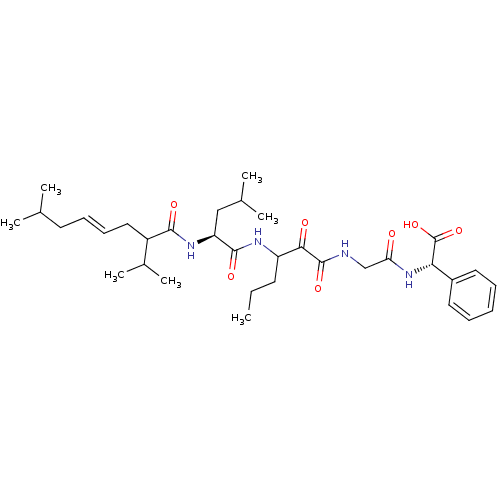
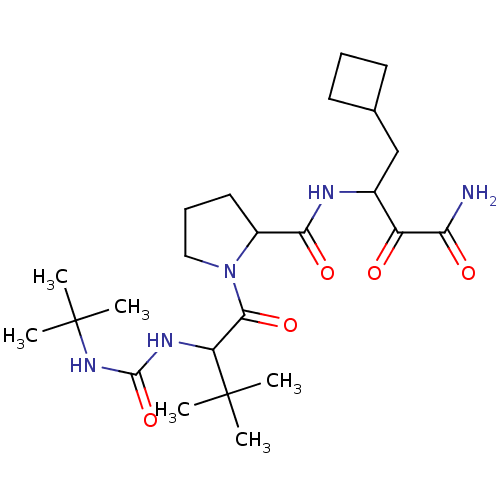
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 10nM ΔG°: -46.4kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
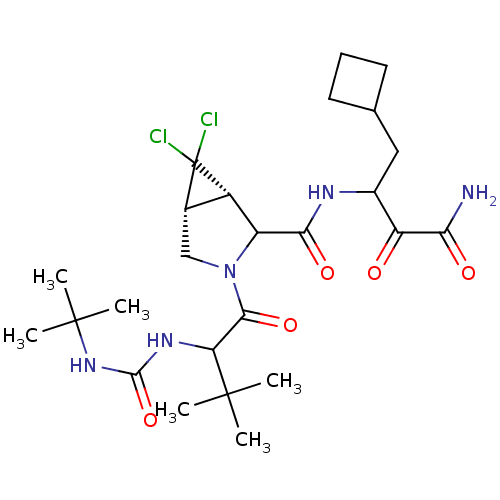
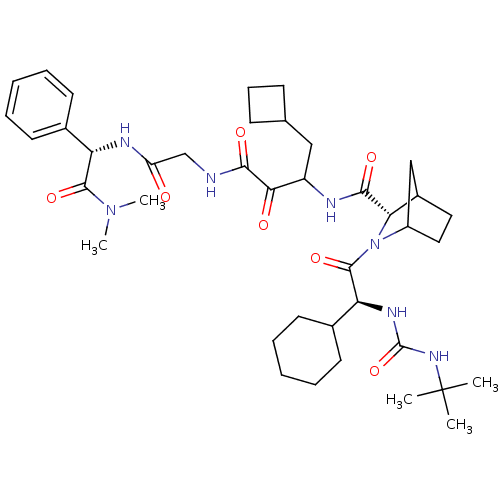
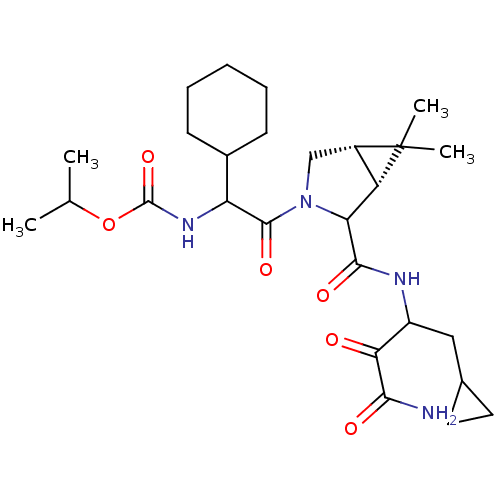
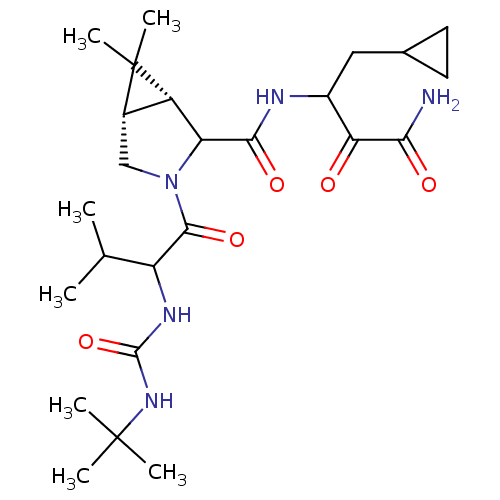
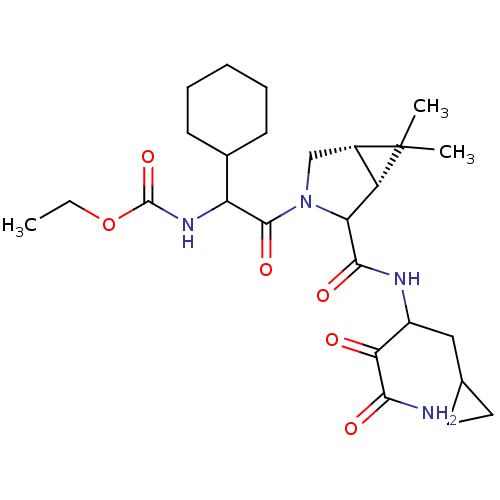
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 13nM ΔG°: -45.8kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
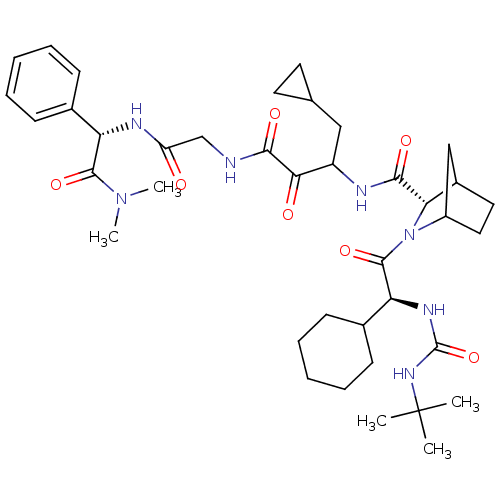
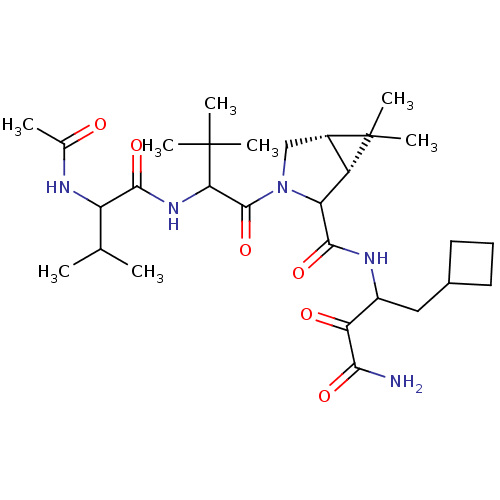
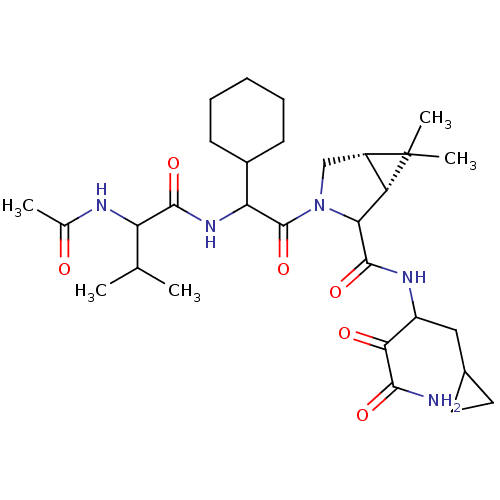
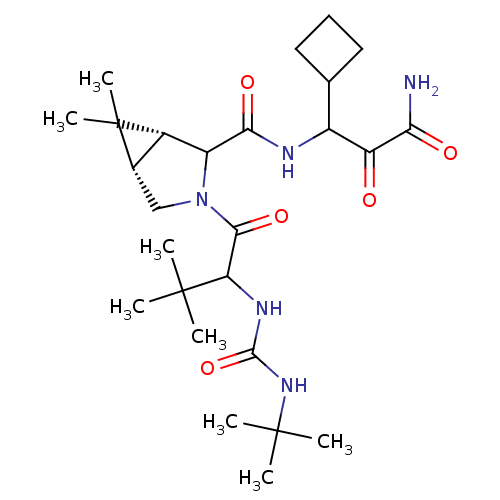
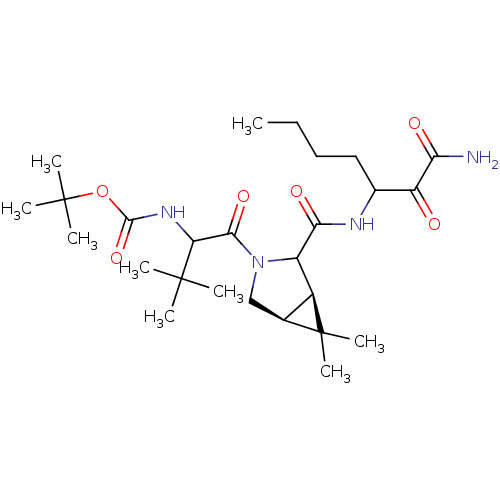
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 14nM ΔG°: -45.6kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
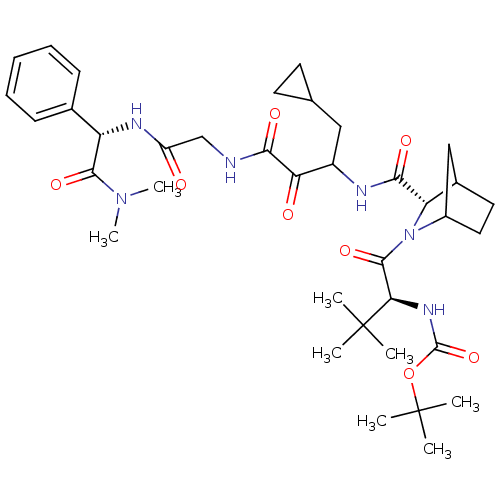
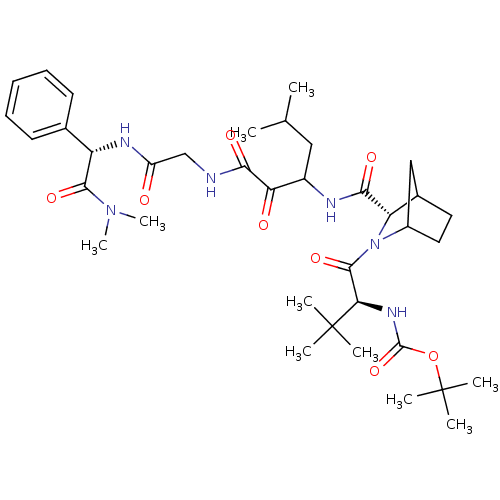
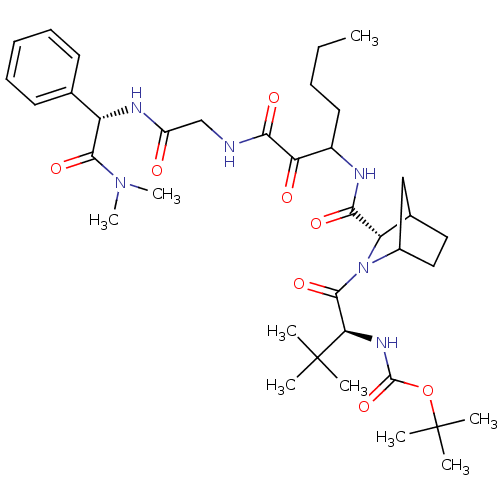
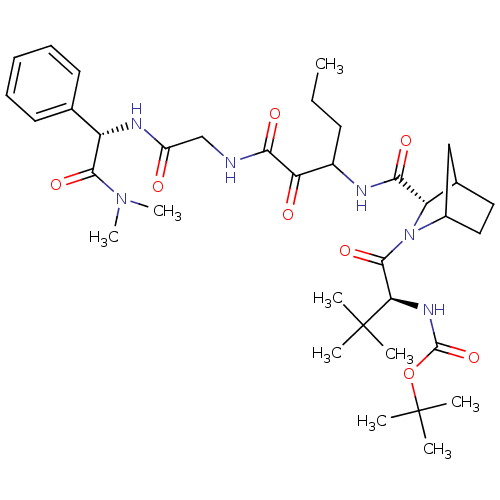
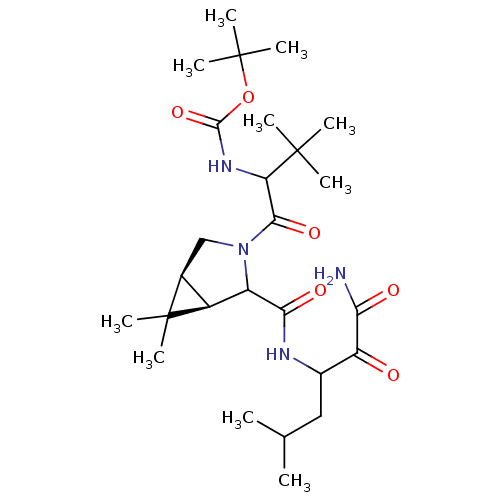
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 15nM ΔG°: -45.4kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 19nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 19nM ΔG°: -44.8kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 19nM ΔG°: -44.8kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 20nM ΔG°: -44.7kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 21nM ΔG°: -44.6kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 27nM ΔG°: -43.9kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 38nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 40nM ΔG°: -42.9kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 41nM ΔG°: -42.9kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 50nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 51nM ΔG°: -42.3kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 53nM ΔG°: -42.2kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 54nM ΔG°: -42.2kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 57nM ΔG°: -42.0kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 60nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 68nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 81nM ΔG°: -41.2kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 90nM ΔG°: -40.9kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 100nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 100nM ΔG°: -40.6kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 110nM ΔG°: -40.4kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
Affinity DataKi: 110nMAssay Description:Ability to inhibit the hydrolysis of chromogenic 4-phenylazophenyl ester from the peptide fragment Ac- DTEDVVP(Nva)-O-4-PAP in a HCV protease continu...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 115nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 140nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
Affinity DataKi: 160nMAssay Description:Ability to inhibit the hydrolysis of chromogenic 4-phenylazophenyl ester from the peptide fragment Ac- DTEDVVP(Nva)-O-4-PAP in a HCV protease continu...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 220nM ΔG°: -38.6kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 230nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
Affinity DataKi: 300nMAssay Description:Ability to inhibit the hydrolysis of chromogenic 4-phenylazophenyl ester from the peptide fragment Ac- DTEDVVP(Nva)-O-4-PAP in a HCV protease continu...More data for this Ligand-Target Pair
Affinity DataKi: 300nMAssay Description:Ability to inhibit the hydrolysis of chromogenic 4-chlorophenylbutyric acid ester from the peptide fragment Ac- DTEDVVP(Nva)-O-4-PAP in a HCV proteas...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 360nM ΔG°: -37.4kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 480nM ΔG°: -36.7kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 500nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
Affinity DataKi: 800nMAssay Description:Ability to inhibit the hydrolysis of chromogenic 4-phenylazophenyl ester from the peptide fragment Ac- DTEDVVP(Nva)-O-4-PAP in a HCV protease continu...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 800nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 960nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 1.00E+3nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 1.40E+3nM ΔG°: -34.0kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 1.50E+3nM ΔG°: -33.8kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 1.60E+3nM ΔG°: -33.6kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 1.80E+3nM ΔG°: -33.3kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
Affinity DataKi: 2.90E+3nMAssay Description:Binding constant towards Human neutrophil elastase HNE proteaseMore data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: >3.30E+3nM ΔG°: >-31.8kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 3.80E+3nM ΔG°: -31.5kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 4.30E+3nM ΔG°: -31.1kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 5.00E+3nMAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair
TargetGenome polyprotein(Hepatitis C virus (HCV genotype 1a, isolate H))
Schering-Plough Research Institute
Schering-Plough Research Institute
Affinity DataKi: 5.50E+3nM ΔG°: -30.5kJ/molepH: 6.5 T: 2°CAssay Description:Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I...More data for this Ligand-Target Pair


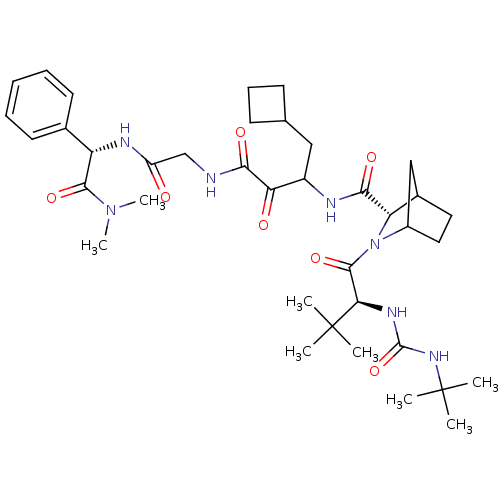
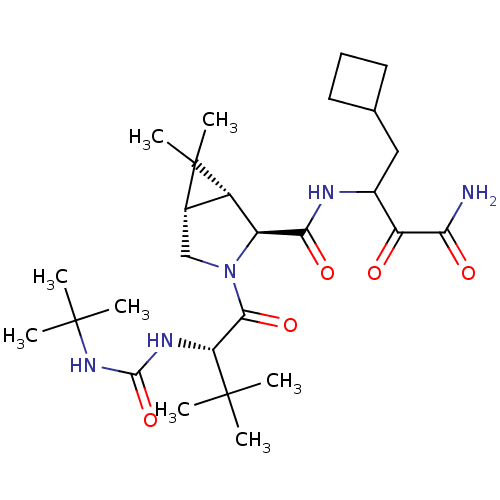
 3D Structure (crystal)
3D Structure (crystal)