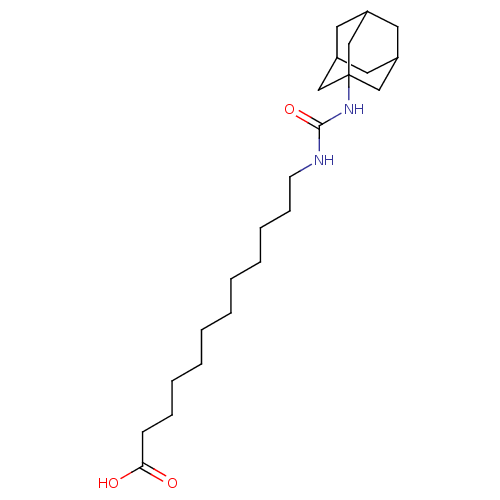
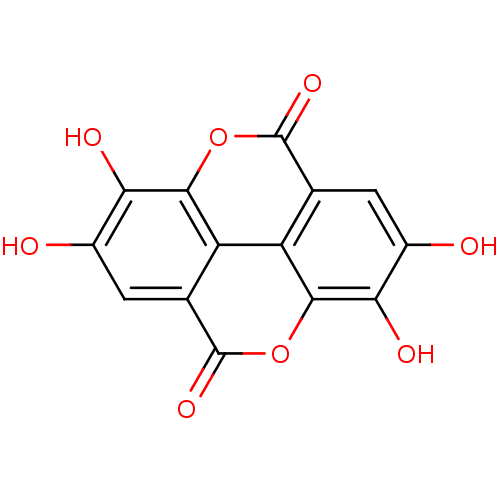
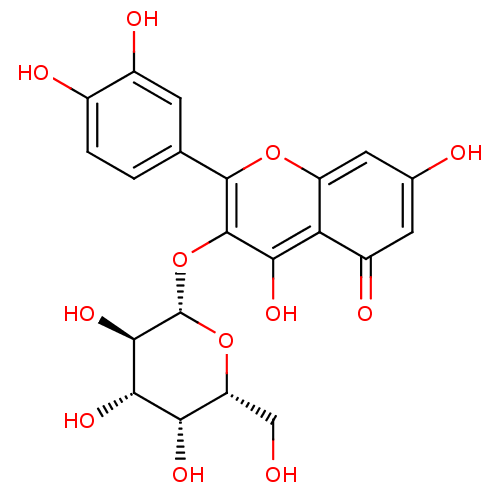
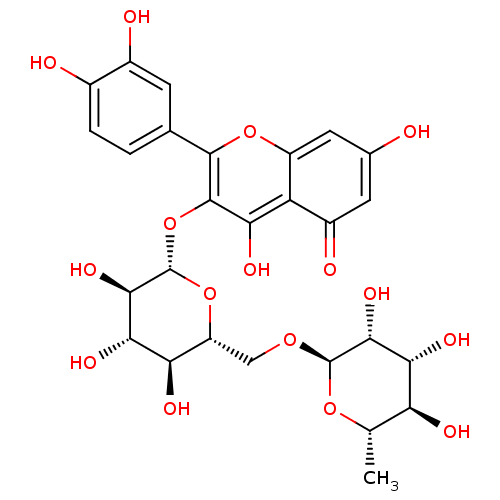
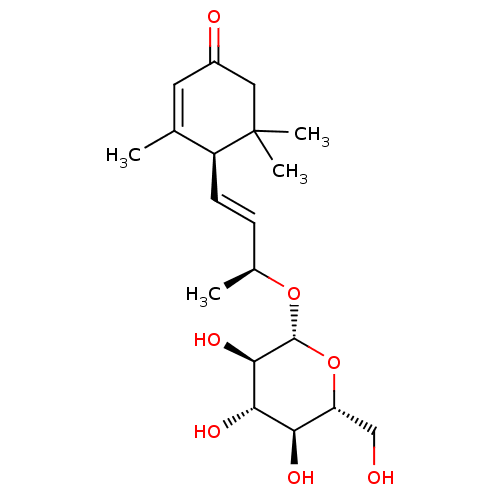
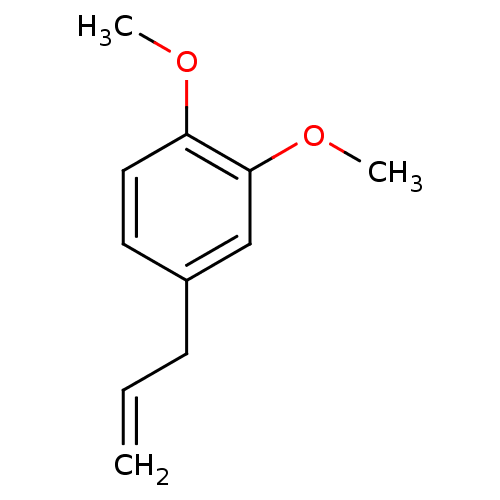
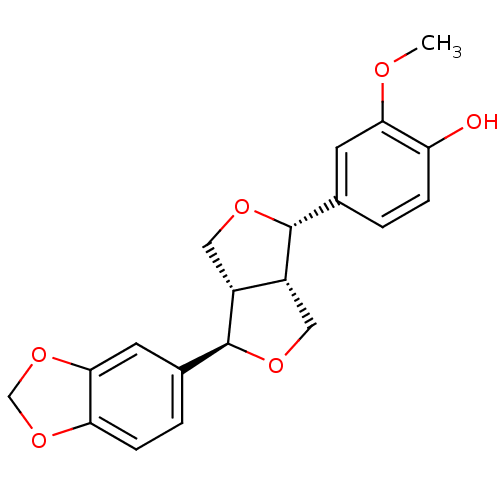
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 10nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 3.89E+3nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
Catholic University of Daegu
Curated by ChEMBL
Catholic University of Daegu
Curated by ChEMBL
Affinity DataIC50: 1.69E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-tyrosine as substrate measured every 1 min for 20 mins by spectrophotometric assayMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.72E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.81E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.86E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 2.07E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 2.18E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 2.38E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 2.43E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 2.74E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 2.84E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 3.35E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 3.61E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetPolyphenol oxidase 2(Agaricus bisporus (Common mushroom))
Catholic University of Daegu
Curated by ChEMBL
Catholic University of Daegu
Curated by ChEMBL
Affinity DataIC50: 3.64E+4nMAssay Description:Inhibition of mushroom tyrosinase using L-tyrosine as substrate measured every 1 min for 20 mins by spectrophotometric assayMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 4.17E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 4.74E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 5.00E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetBifunctional epoxide hydrolase 2(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 5.00E+4nMAssay Description:Inhibition of sEH (unknown origin) assessed as substrate PHOME hydrolysis after 1 hr by fluorescence methodMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 5.90E+4nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 8.52E+4nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 8.63E+4nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 1.01E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 1.12E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 1.12E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 1.16E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 1.46E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 1.52E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 1.75E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 1.94E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 2.02E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 2.50E+5nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 3.10E+5nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
National Institute of Medicinal Materials
Curated by ChEMBL
National Institute of Medicinal Materials
Curated by ChEMBL
Affinity DataIC50: 3.72E+5nMAssay Description:Inhibition of xanthine oxidase- mediated uric acid formation after 5 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 4.00E+5nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 4.30E+5nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 4.80E+5nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 5.20E+5nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 6.50E+5nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of rat intestinal sucrase using p-nitrophenyl-alpha-d-glucopyranoside as substrate incubated for 10 mins prior to substrate addition measu...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataEC50: 4.00E+4nMAssay Description:Transactivation of GAL4-fused PPARgamma LBD expressed in HepG2 cells after 20 hrs by luminescence assayMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataEC50: >6.00E+4nMAssay Description:Transactivation of GAL4-fused PPARalpha LBD expressed in HepG2 cells after 20 hrs by luminescence assayMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataEC50: >6.00E+4nMAssay Description:Transactivation of GAL4-fused PPARalpha LBD expressed in HepG2 cells after 20 hrs by luminescence assayMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha(Homo sapiens (Human))
Chungnam National University
Curated by ChEMBL
Chungnam National University
Curated by ChEMBL
Affinity DataEC50: >6.00E+4nMAssay Description:Transactivation of GAL4-fused PPARalpha LBD expressed in HepG2 cells after 20 hrs by luminescence assayMore data for this Ligand-Target Pair