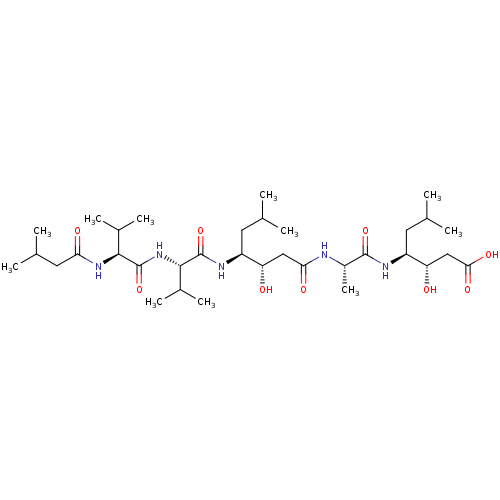
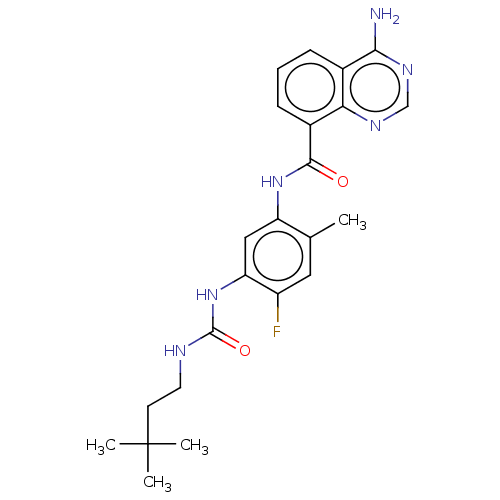
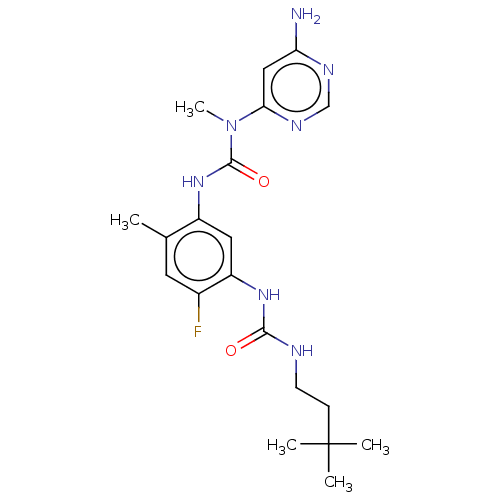
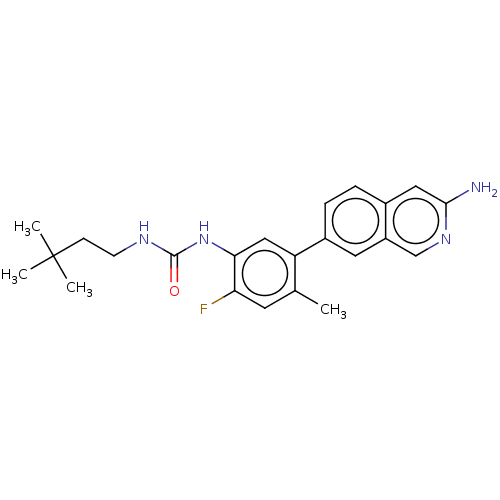
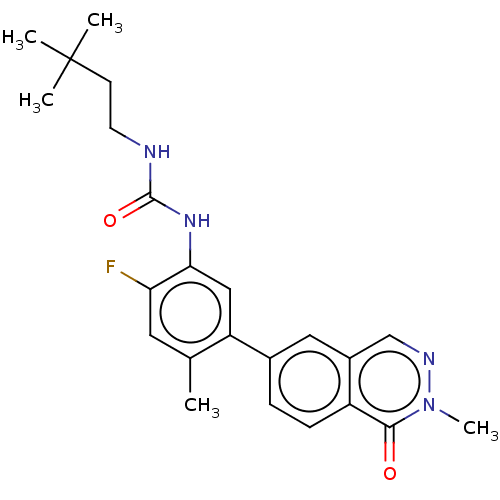
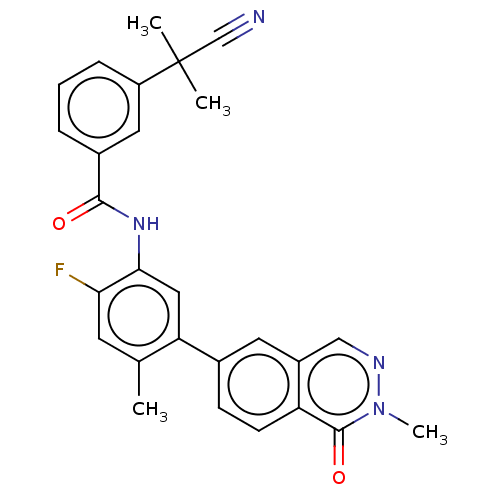
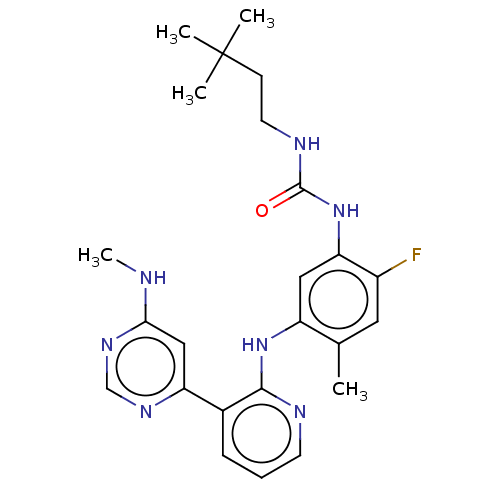
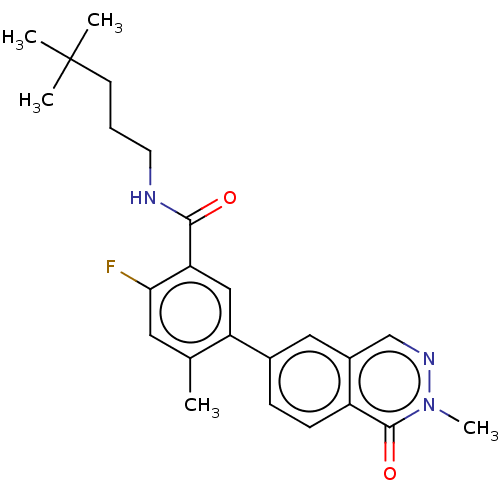
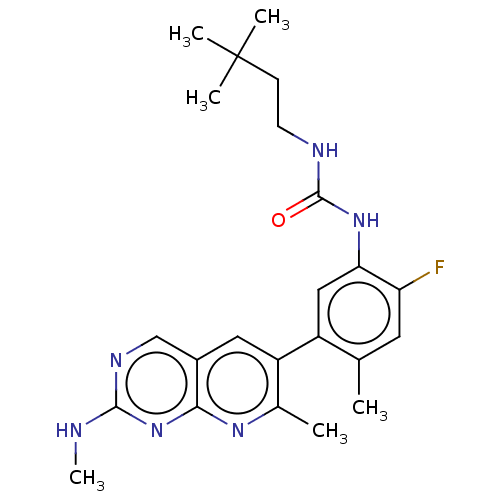
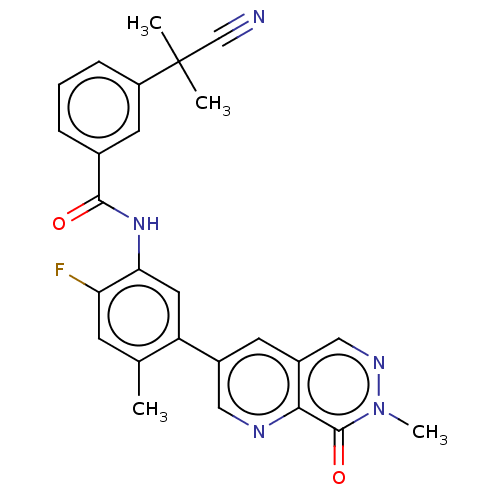
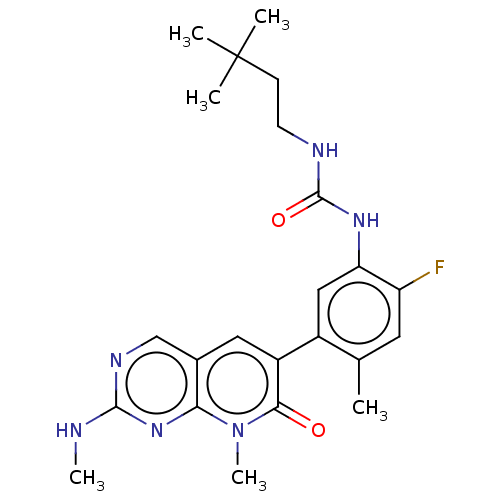
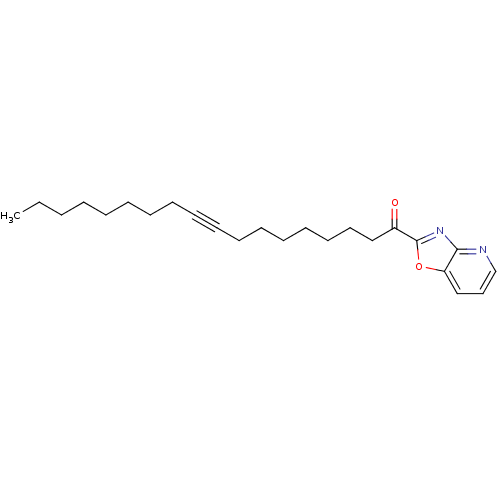
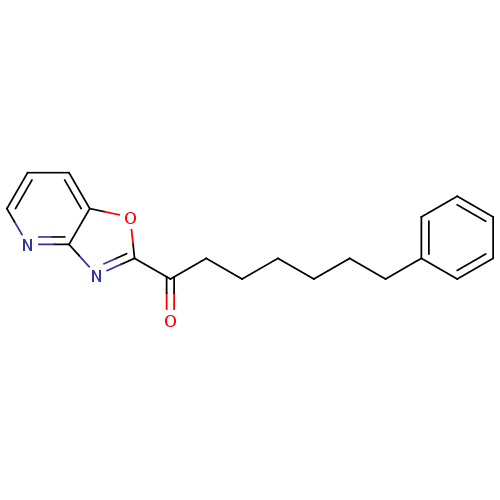
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
University of Queensland
Curated by ChEMBL
University of Queensland
Curated by ChEMBL
Affinity DataKi: 0.00400nMAssay Description:Binding affinity against ritonavir-resistant strains.Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.0100nMAssay Description:Binding affinity against human cathepsin DChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.0130nMAssay Description:Binding affinity against Elastase.Checked by AuthorMore data for this Ligand-Target Pair
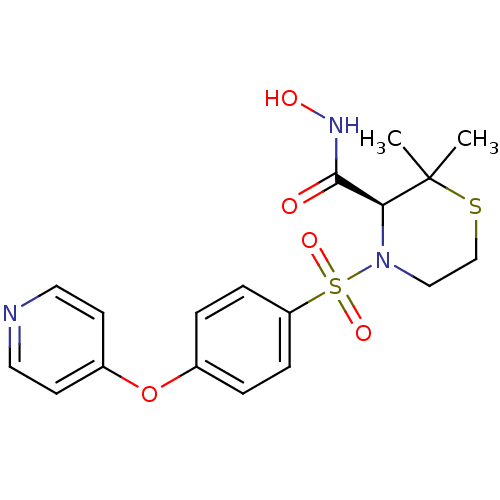
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
University of Queensland
Curated by ChEMBL
University of Queensland
Curated by ChEMBL
Affinity DataKi: 0.0310nMAssay Description:Binding affinity against HIV-1 protease .Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.0380nMAssay Description:Inhibition of matrix metalloprotease-13 (MMP-13).Checked by AuthorMore data for this Ligand-Target Pair
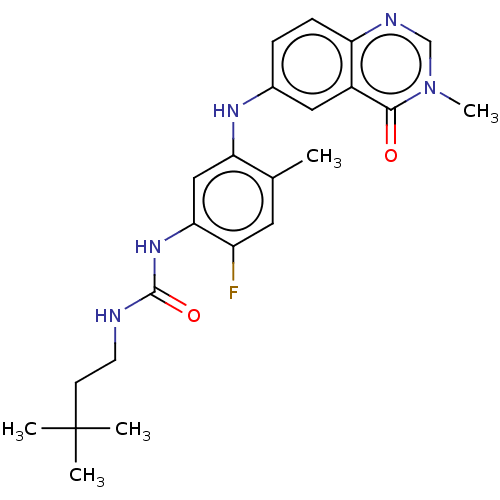
Affinity DataKi: <0.0500nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) using inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA a...More data for this Ligand-Target Pair
Affinity DataKi: <0.0500nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) using inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA a...More data for this Ligand-Target Pair
Affinity DataKi: <0.0500nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) using inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA a...More data for this Ligand-Target Pair
Affinity DataKi: <0.0500nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) using inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA a...More data for this Ligand-Target Pair
Affinity DataKi: <0.0500nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured a...More data for this Ligand-Target Pair
Affinity DataKi: <0.0500nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured a...More data for this Ligand-Target Pair
Affinity DataKi: <0.0500nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0560nMAssay Description:Binding affinity against Thrombin.Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.0600nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) using inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0600nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0610nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) using inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0610nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) using inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0610nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0610nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0620nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured a...More data for this Ligand-Target Pair
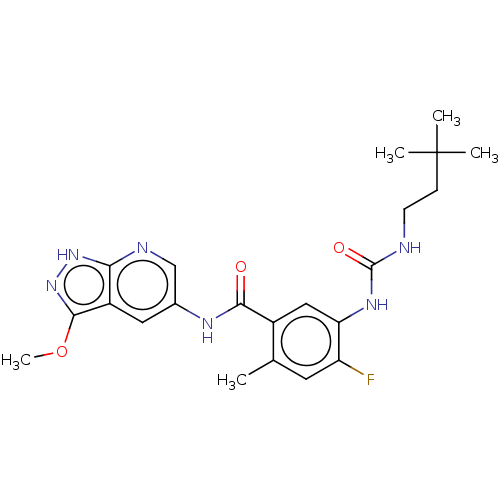
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA assayMore data for this Ligand-Target Pair
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA assayMore data for this Ligand-Target Pair
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA assayMore data for this Ligand-Target Pair
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA assayMore data for this Ligand-Target Pair
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA assayMore data for this Ligand-Target Pair
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured after 9...More data for this Ligand-Target Pair
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured after 9...More data for this Ligand-Target Pair
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured after 9...More data for this Ligand-Target Pair
Affinity DataKi: <0.0800nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured after 9...More data for this Ligand-Target Pair
Affinity DataKi: 0.0810nMAssay Description:Inhibition of CRAF Y340D/Y341D mutant (unknown origin) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0830nMAssay Description:Inhibition of matrix metalloprotease-2 (MMP-2).Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Binding affinity against Coagulation factor XChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.102nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured after 9...More data for this Ligand-Target Pair
Affinity DataKi: 0.120nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured after 9...More data for this Ligand-Target Pair
Affinity DataKi: 0.134nMAssay Description:Binding affinity against HIV-1 protease .Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.139nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.139nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured after 9...More data for this Ligand-Target Pair
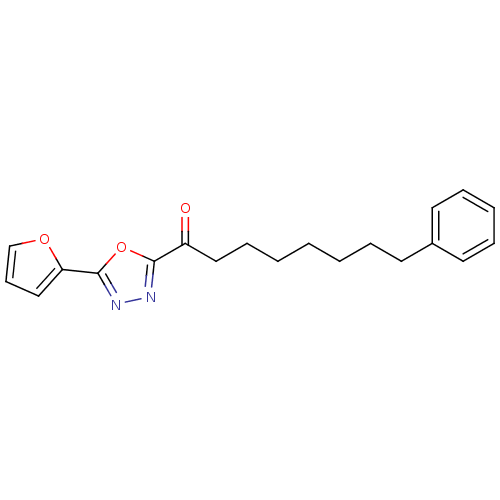
TargetFatty-acid amide hydrolase 1 [30-579](Rattus norvegicus (rat))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.140nMAssay Description:Inhibitory constant determined against recombinant Fatty-acid amide hydrolase from rat expressed in Escherichia coliMore data for this Ligand-Target Pair
Affinity DataKi: 0.165nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) inactive MAP2K1 as substrate preincubated for 30 mins measured after 90 mins by DELFIA assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.190nMAssay Description:Inhibition of BRAF (unknown origin) (416 to 766) assessed as using inactive phosphorylated MAP2K1 substrate preincubated for 30 mins measured after 9...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.200nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1 [30-579](Rattus norvegicus (rat))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.200nMAssay Description:Inhibitory constant determined against recombinant Fatty-acid amide hydrolase from rat expressed in Escherichia coliMore data for this Ligand-Target Pair
Affinity DataKi: 0.210nMAssay Description:Binding affinity against Thrombin.Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.220nMAssay Description:Inhibition of matrix metalloprotease-2 (MMP-2).Checked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 0.270nMAssay Description:Inhibition of matrix metalloprotease-3 (MMP-3).Checked by AuthorMore data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.280nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1 [30-579](Rattus norvegicus (rat))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.280nMAssay Description:Inhibitory constant determined against recombinant Fatty-acid amide hydrolase from rat expressed in Escherichia coliMore data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.290nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
Affinity DataKi: 0.300nMAssay Description:Binding affinity against Thrombin.Checked by AuthorMore data for this Ligand-Target Pair
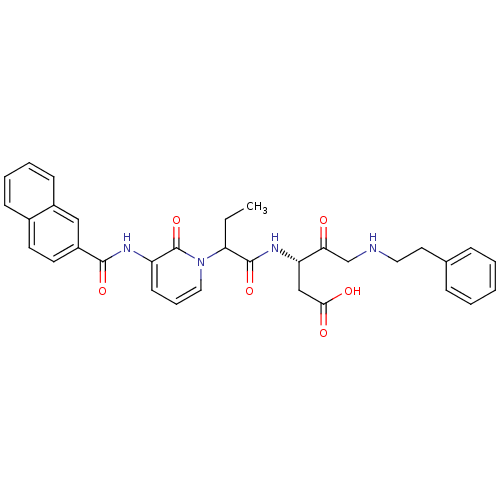
Affinity DataKi: 0.370nMAssay Description:The binding affinity against IL-1 beta converting enzymeChecked by AuthorMore data for this Ligand-Target Pair


 3D Structure (crystal)
3D Structure (crystal)