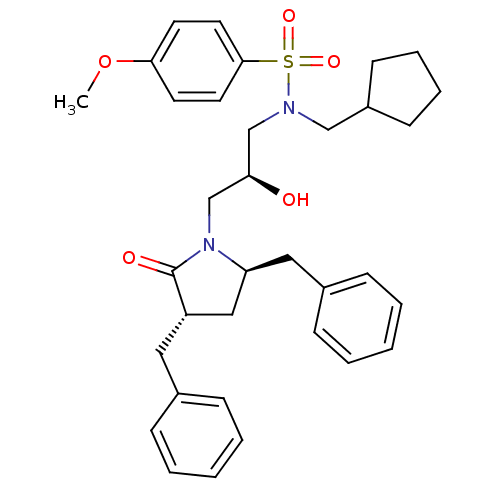
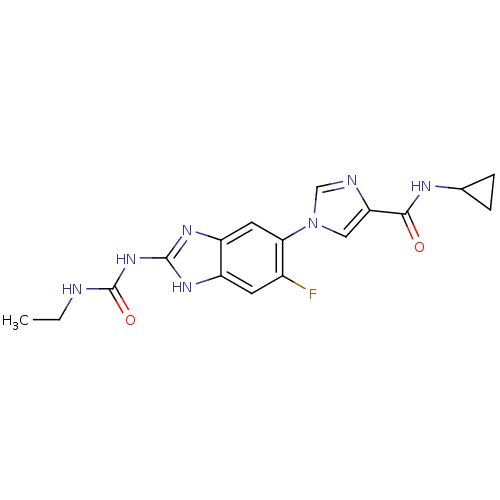
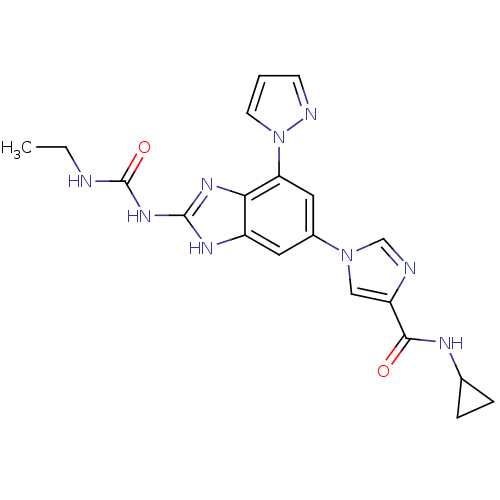
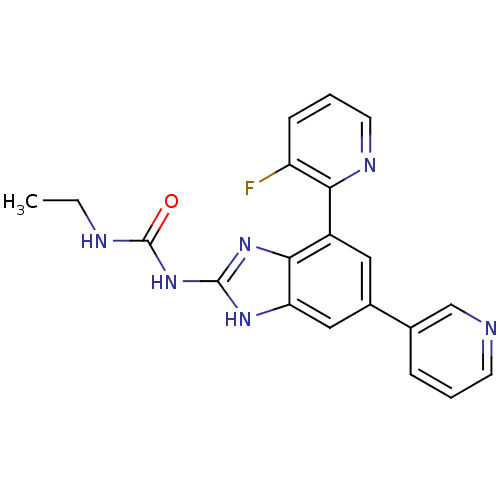
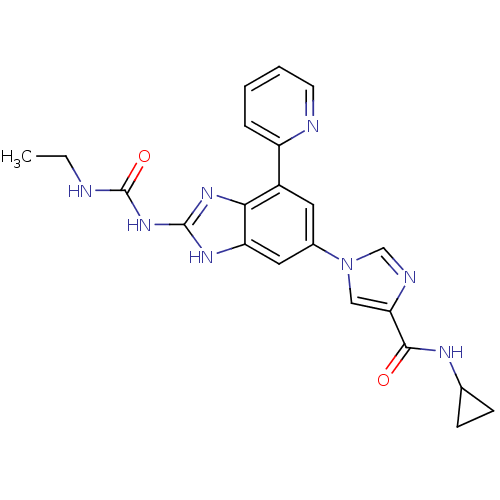
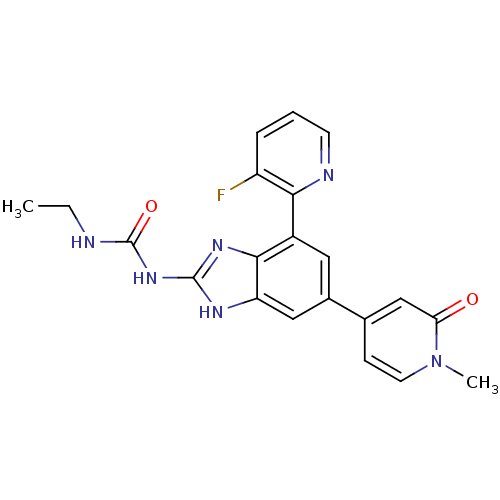
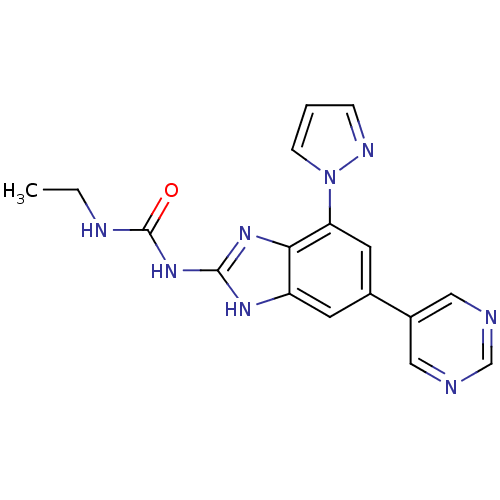
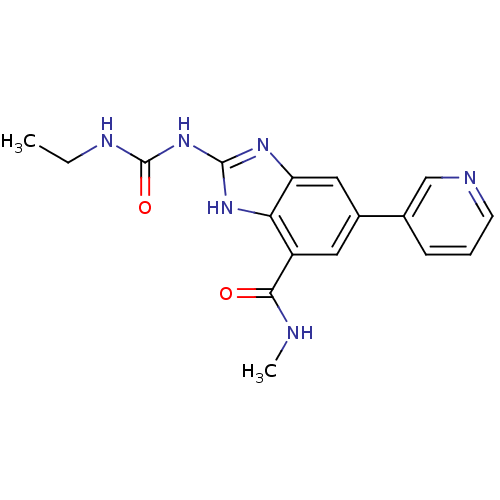
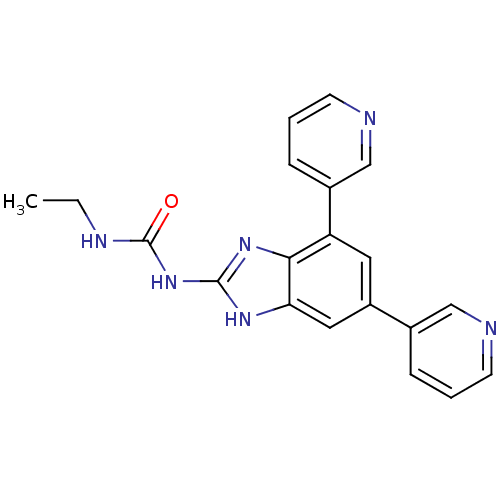
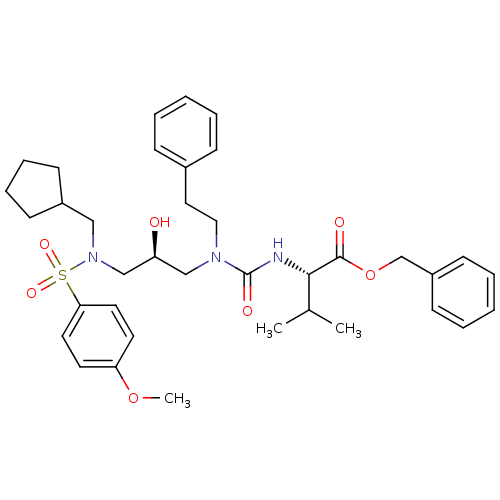
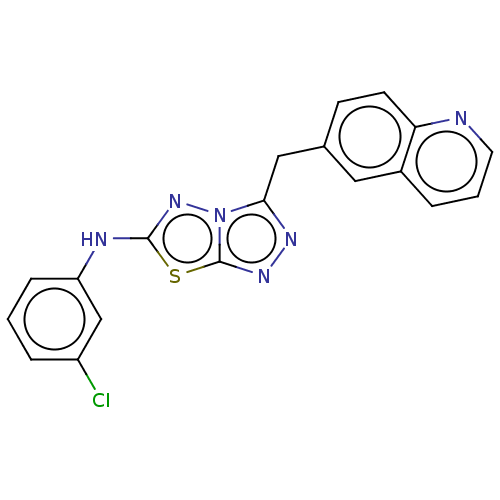
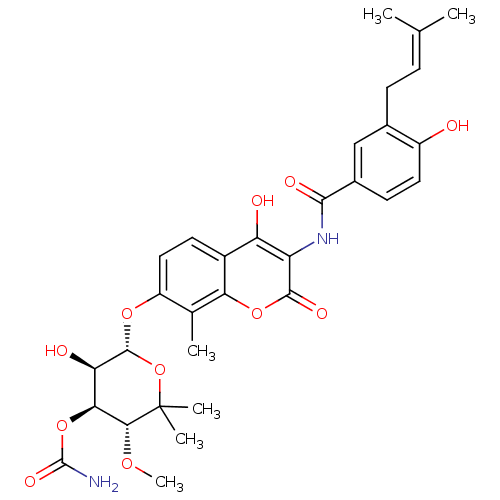
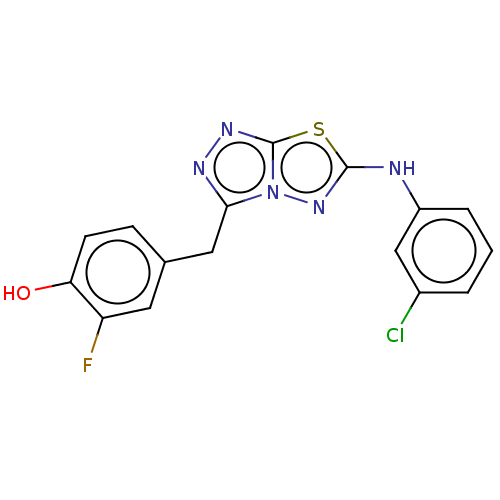
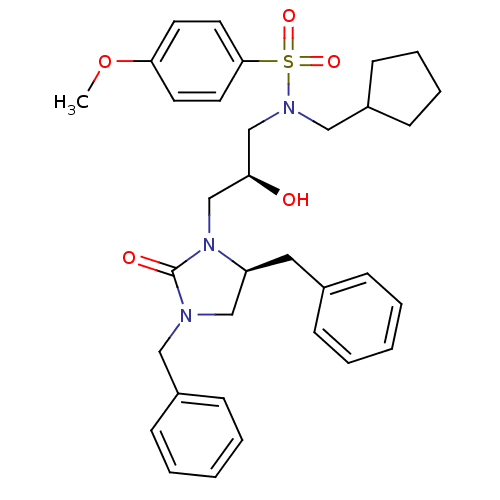
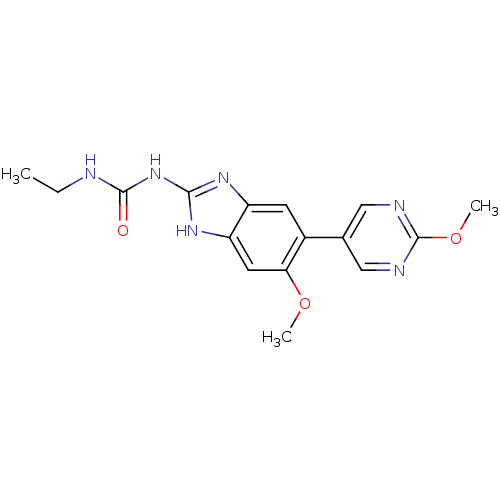
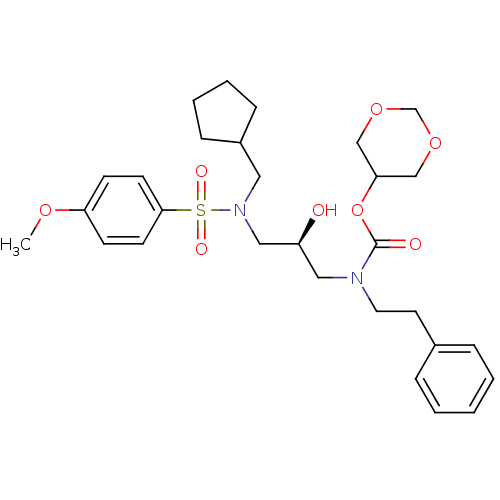
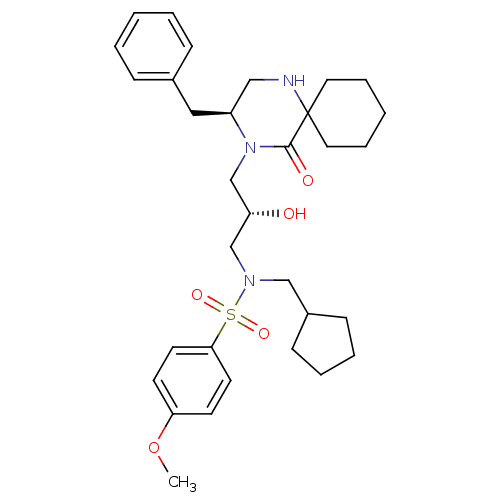
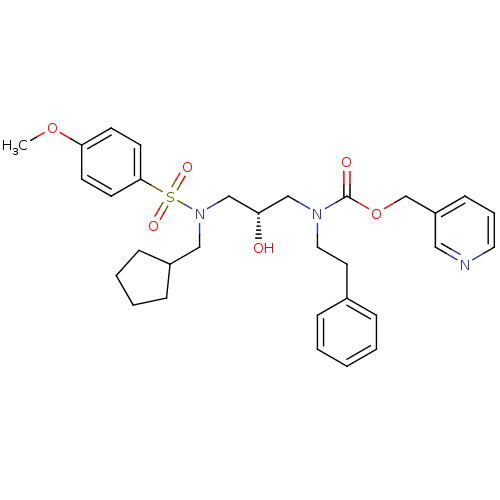
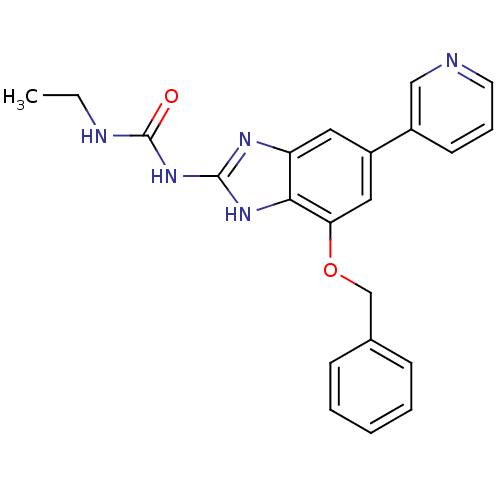
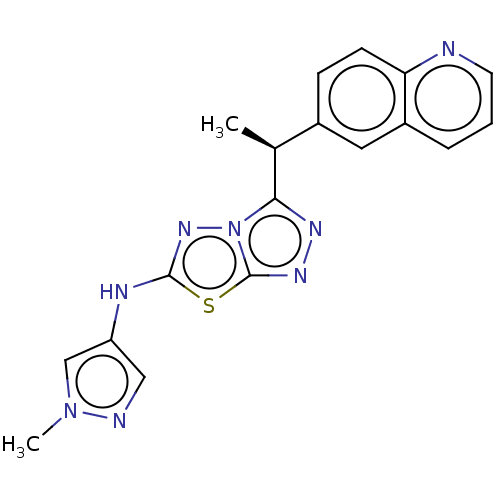
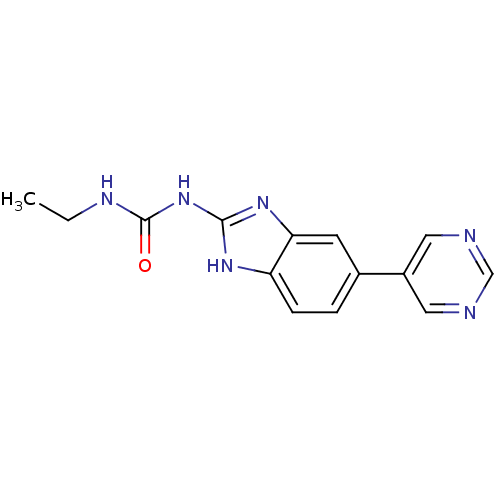
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: <0.100nMAssay Description:Binding affinity to HIV-1 protease was determinedMore data for this Ligand-Target Pair
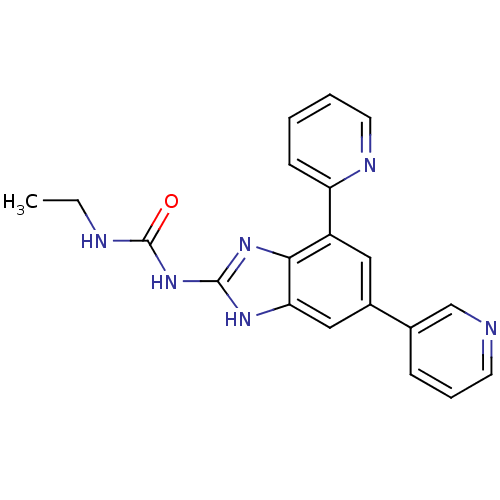
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 0.5nMAssay Description:Compound was assayed for inhibition against HIV protease activityMore data for this Ligand-Target Pair
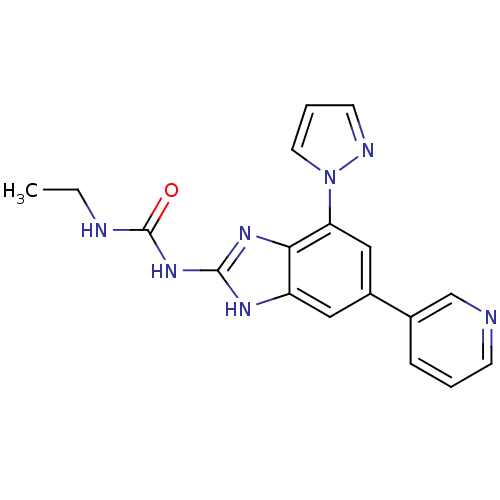
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 0.600nMAssay Description:Compound was assayed for inhibition against HIV protease activityMore data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 5nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 6nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 7nMAssay Description:Binding affinity to HIV-1 protease was determinedMore data for this Ligand-Target Pair
Affinity DataKi: 7nM ΔG°: -47.3kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 7nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 7nMAssay Description:Inhibition of c-Met (unknown origin) using poly Glu-Tyr as substrate preincubated for 10 mins followed by ATP addition by phosphoenolpyruvate/pyruvat...More data for this Ligand-Target Pair
Affinity DataKi: 8nMAssay Description:Inhibition of c-Met (unknown origin) using poly Glu-Tyr as substrate preincubated for 10 mins followed by ATP addition by phosphoenolpyruvate/pyruvat...More data for this Ligand-Target Pair
Affinity DataKi: 8nM ΔG°: -47.0kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nM ΔG°: -46.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nM ΔG°: -46.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nM ΔG°: -46.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nMAssay Description:Inhibition of c-Met (unknown origin) using poly Glu-Tyr as substrate preincubated for 10 mins followed by ATP addition by phosphoenolpyruvate/pyruvat...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 12nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 12nM ΔG°: -46.0kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 14nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 14nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 14nM ΔG°: -45.6kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 15nMAssay Description:Compound was assayed for inhibition against HIV protease activityMore data for this Ligand-Target Pair
Affinity DataKi: 15nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 15nM ΔG°: -45.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 15nMAssay Description:Binding affinity to HIV-1 protease was determinedMore data for this Ligand-Target Pair
Affinity DataKi: 15nMAssay Description:Inhibition of c-Met (unknown origin) using poly Glu-Tyr as substrate preincubated for 10 mins followed by ATP addition by phosphoenolpyruvate/pyruvat...More data for this Ligand-Target Pair
Affinity DataKi: 16nM ΔG°: -45.2kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 17nMAssay Description:Compound was assayed for inhibition against HIV protease activityMore data for this Ligand-Target Pair
Affinity DataKi: 17nM ΔG°: -45.1kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 17nMAssay Description:Compound was assayed for inhibition against HIV protease activityMore data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 17nMAssay Description:Binding affinity to HIV-1 protease was determinedMore data for this Ligand-Target Pair
Affinity DataKi: 18nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 20nMAssay Description:Inhibition of c-Met (unknown origin) using poly Glu-Tyr as substrate preincubated for 10 mins followed by ATP addition by phosphoenolpyruvate/pyruvat...More data for this Ligand-Target Pair
Affinity DataKi: 20nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 20nMAssay Description:Binding affinity to HIV-1 protease was determinedMore data for this Ligand-Target Pair
Affinity DataKi: 20nMAssay Description:Inhibition of c-Met (unknown origin) using poly Glu-Tyr as substrate preincubated for 10 mins followed by ATP addition by phosphoenolpyruvate/pyruvat...More data for this Ligand-Target Pair
TargetGenome polyprotein/Non-structural protein 4A(Hepatitis C virus)
Vertex Pharmaceuticals Inc.
Curated by ChEMBL
Vertex Pharmaceuticals Inc.
Curated by ChEMBL
Affinity DataKi: 20nMAssay Description:Inhibitory activity against hepatitis C virus NS3.4A proteaseMore data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 20nMAssay Description:Binding affinity to HIV-1 protease was determinedMore data for this Ligand-Target Pair
Affinity DataKi: 21nMAssay Description:Inhibition of c-Met (unknown origin) using poly Glu-Tyr as substrate preincubated for 10 mins followed by ATP addition by phosphoenolpyruvate/pyruvat...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Vertex Pharmaceuticals Inc
Curated by ChEMBL
Affinity DataKi: 22nMAssay Description:Binding affinity of the compound against HIV-1 protease was determinedMore data for this Ligand-Target Pair
Affinity DataKi: 23nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair


 3D Structure (crystal)
3D Structure (crystal)