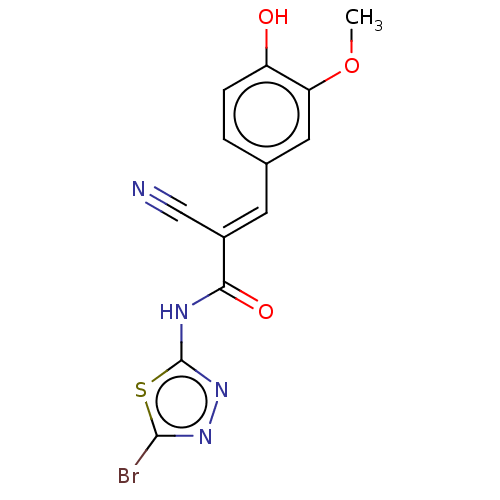
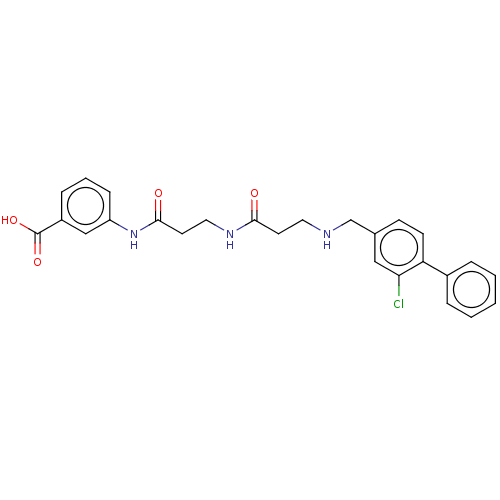
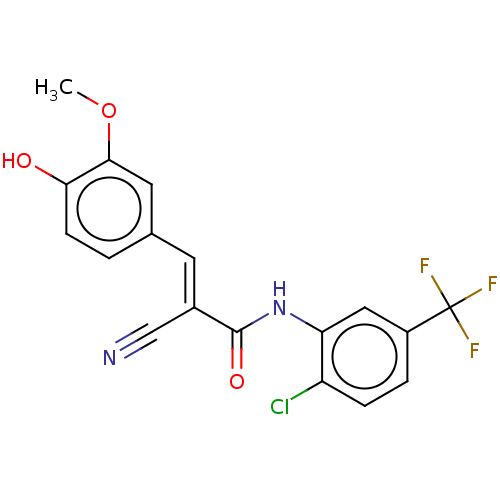
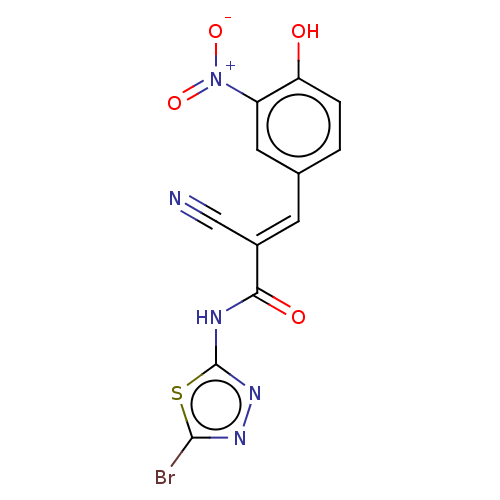
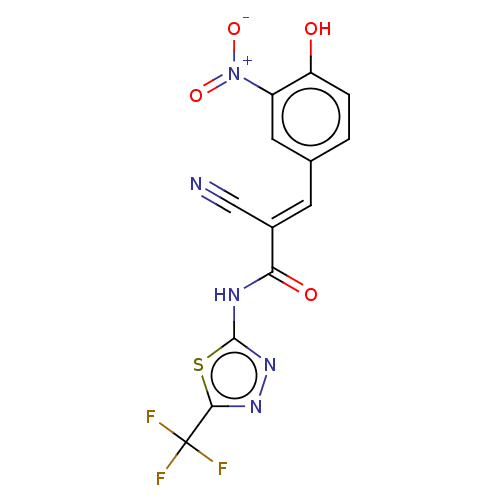
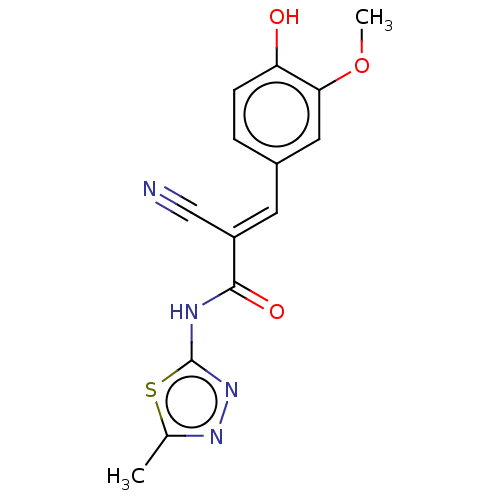
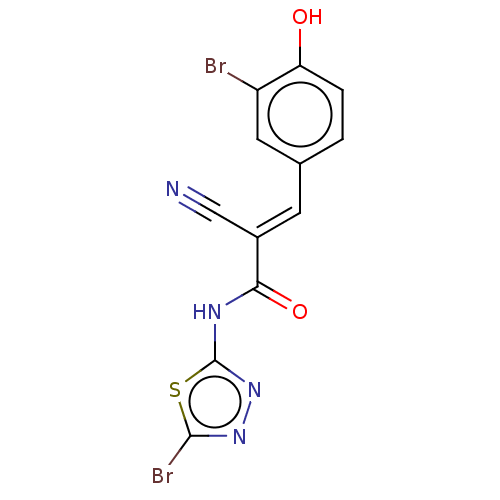
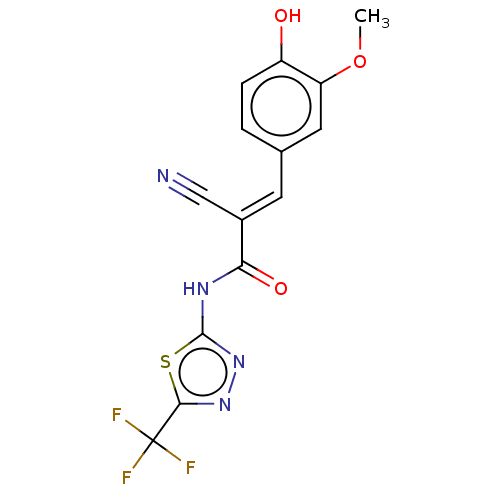
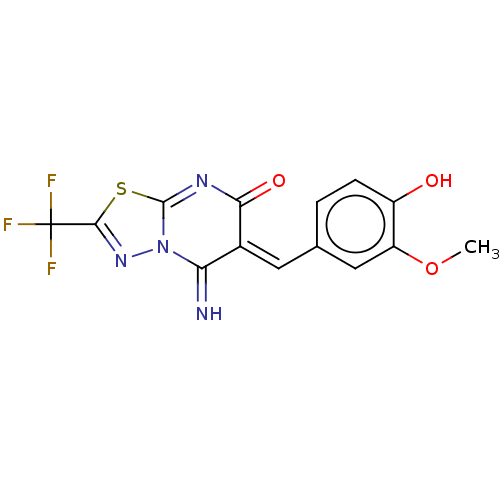
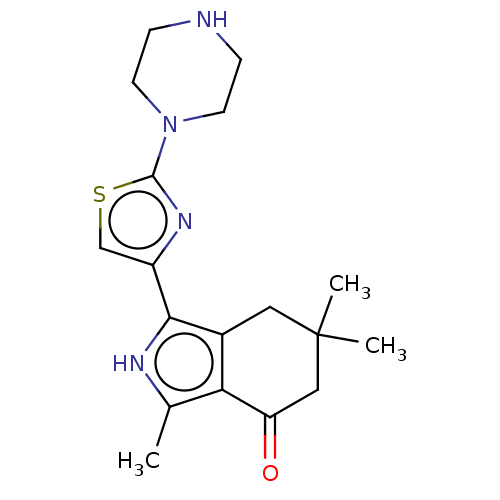
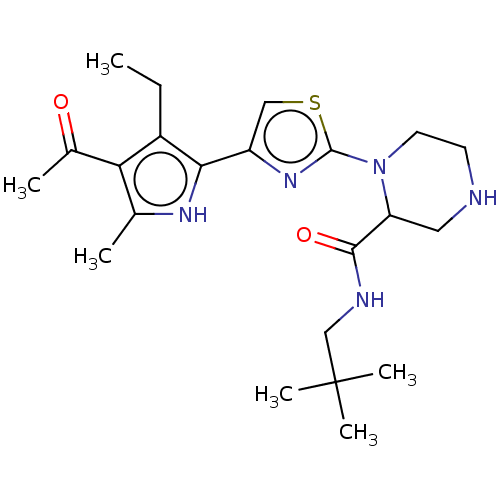
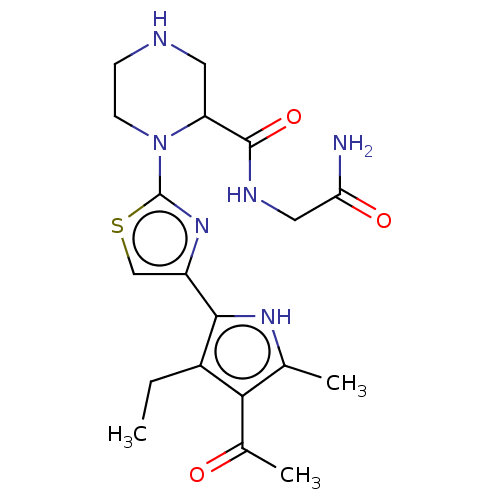
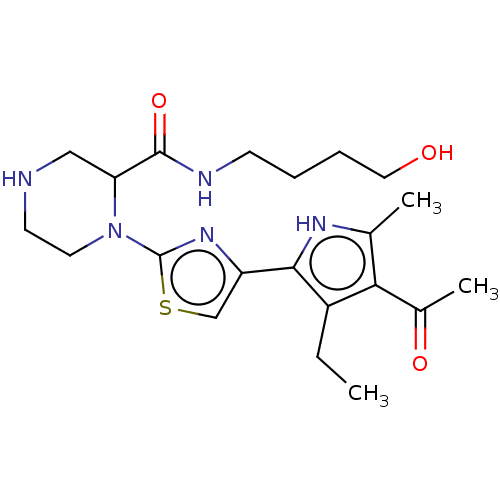
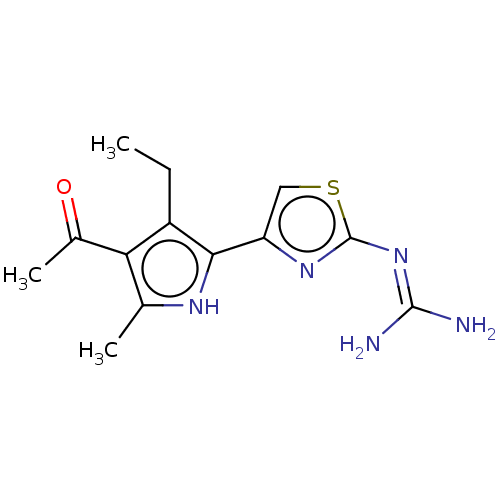
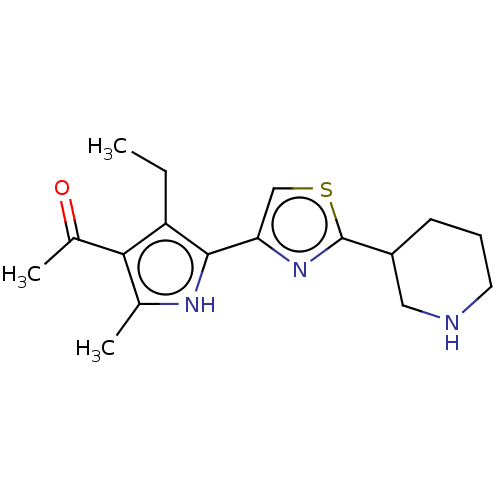
Affinity DataIC50: 1nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
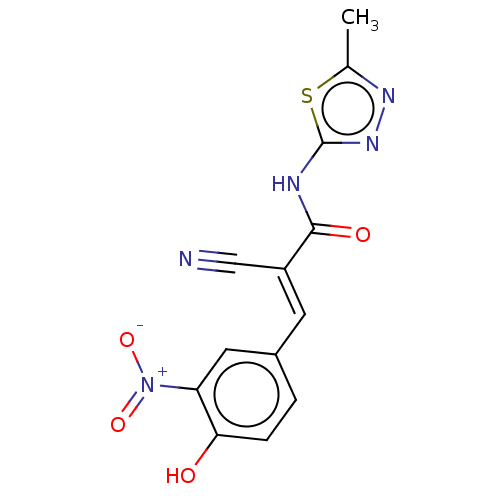
Affinity DataIC50: 280nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
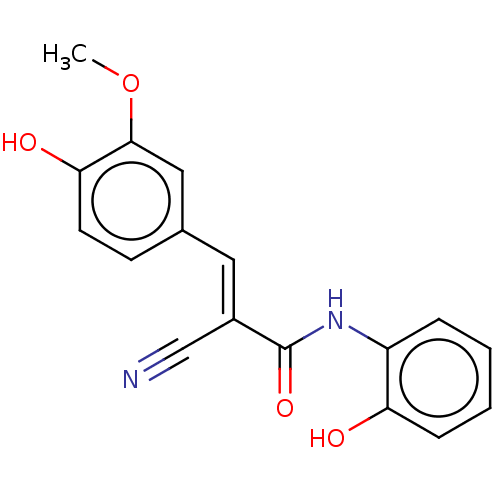
Affinity DataIC50: 320nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
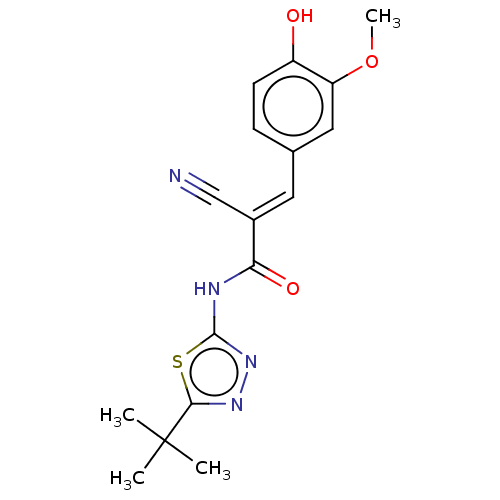
Affinity DataIC50: 330nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 360nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 370nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 370nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 370nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 710nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 880nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.30E+3nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.37E+3nMAssay Description:Inhibition of recombinant human CK2alpha (1 to 336 residues) expressed in Escherichia coli BL21 (DE3) using RRADDSDDDD as substrate incubated for 10 ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.40E+3nMAssay Description:Inhibition of recombinant human SRPK1 expressed in Escherichia coli using RS peptide as substrate incubated for 15 mins in presence of ATP by Kinase-...More data for this Ligand-Target Pair

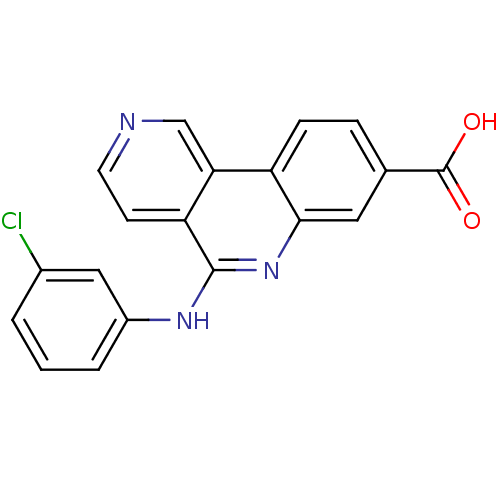
 3D Structure (crystal)
3D Structure (crystal)