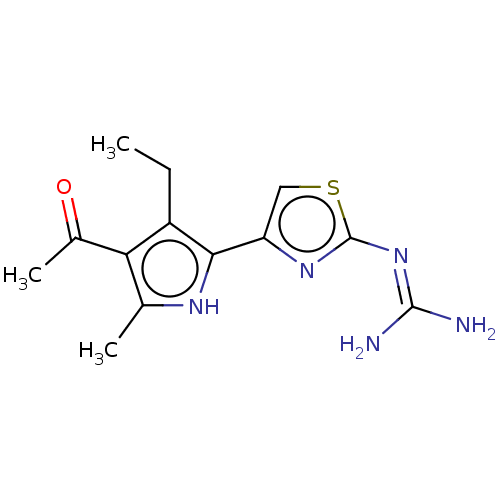
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
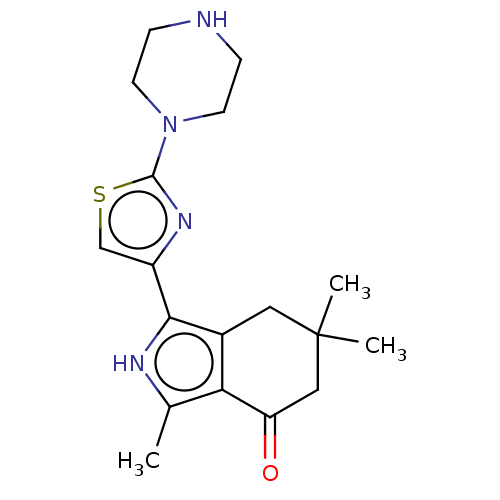
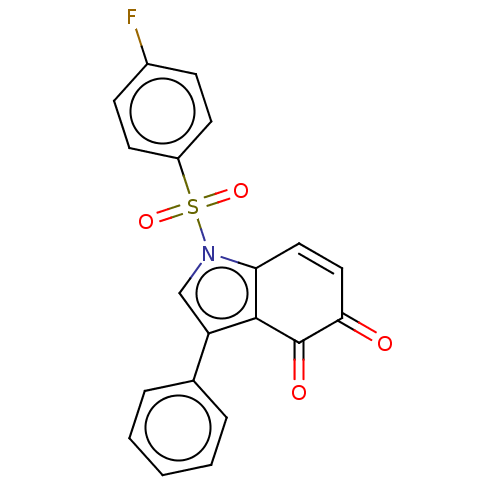
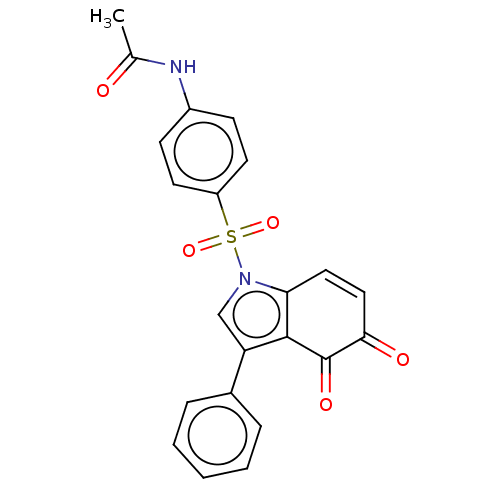
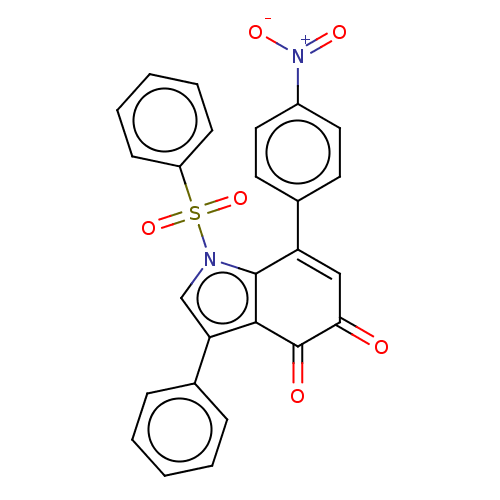
Affinity DataKi: 13nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
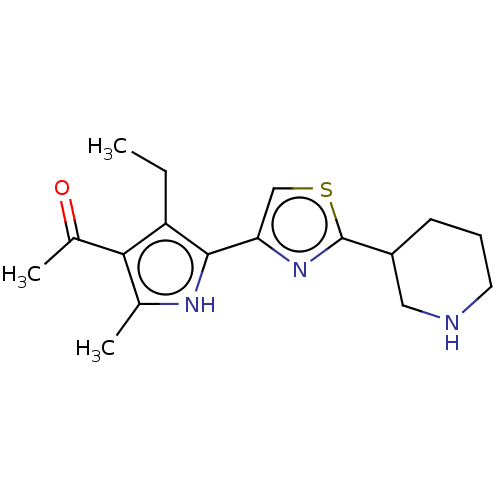
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
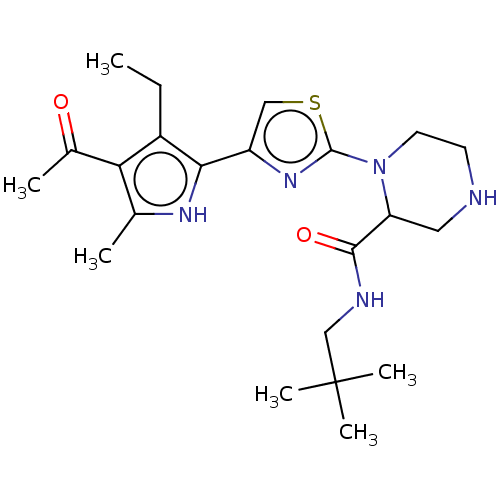
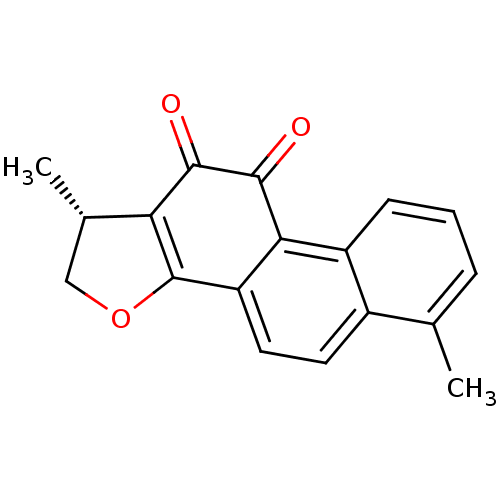
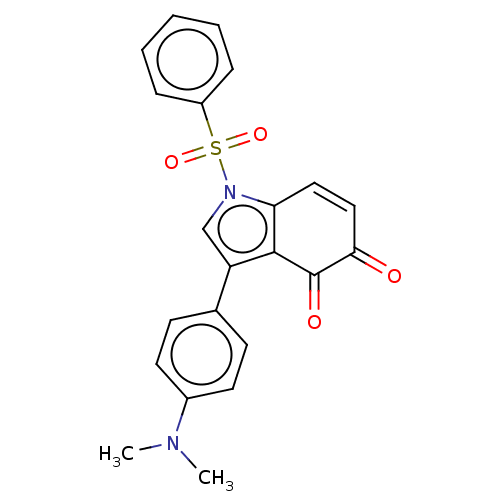
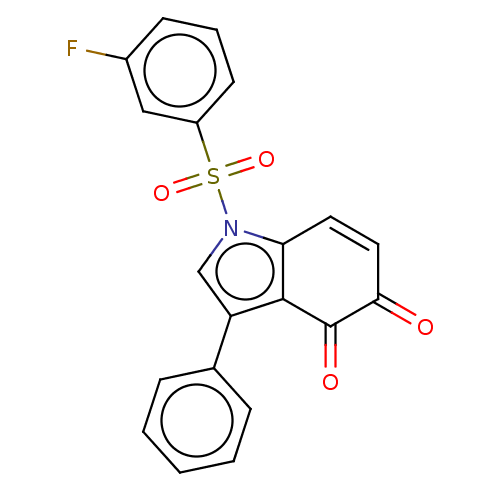
Affinity DataKi: 15nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
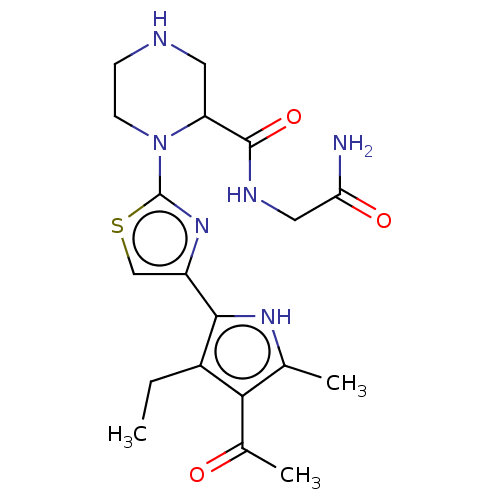
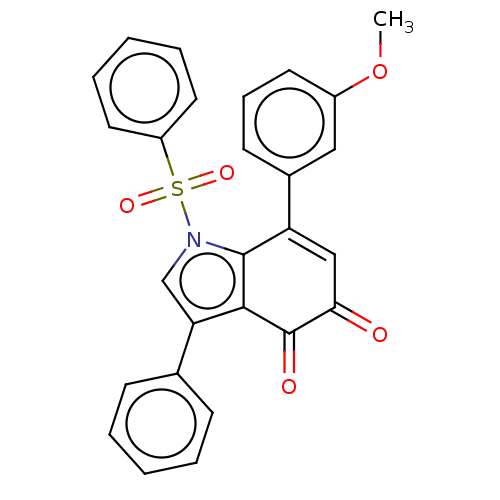
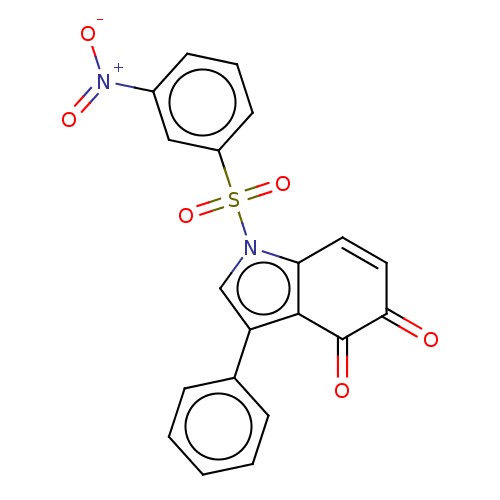
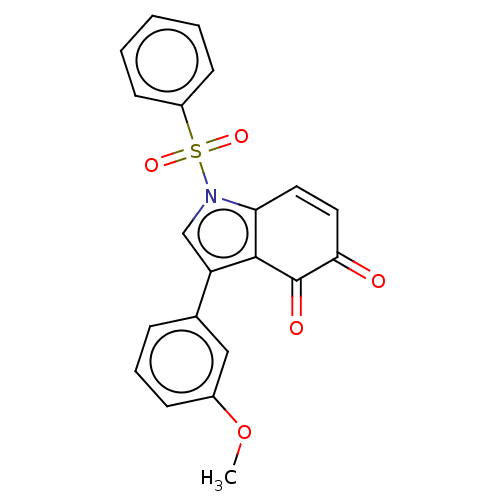
Affinity DataKi: 41nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
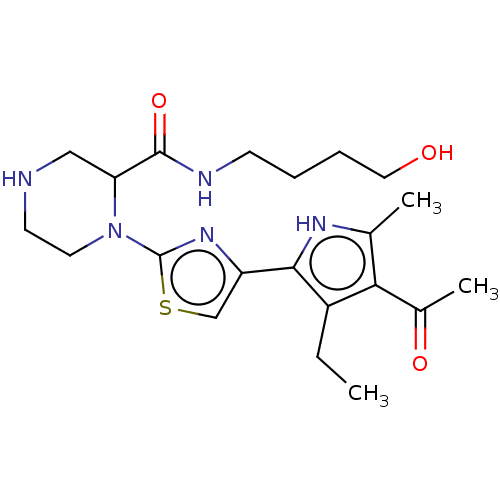
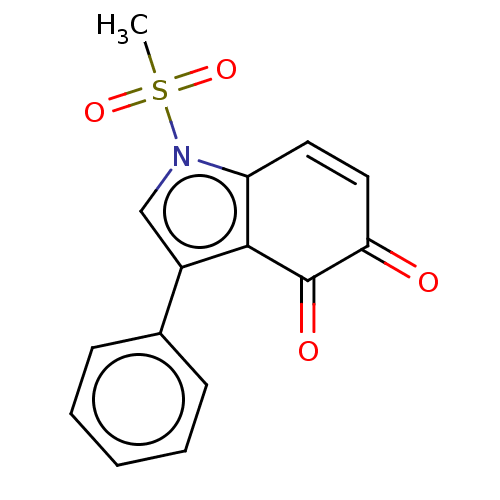
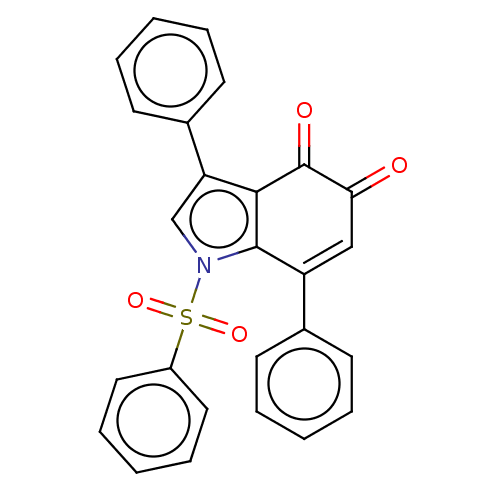
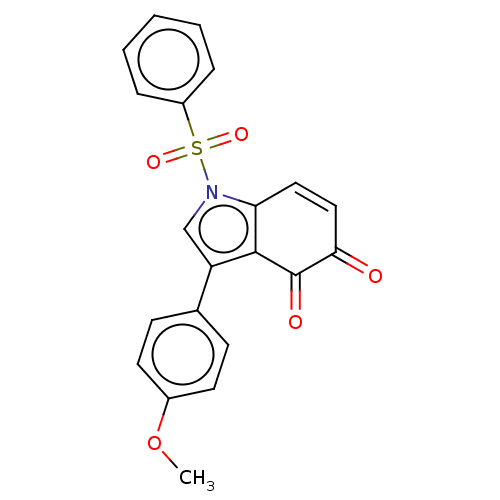
Affinity DataKi: 48nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: 50nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: 55nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: 56nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: 81nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >100nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >100nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >100nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >100nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >100nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >200nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >200nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >200nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair
TargetELAV-like protein 1(Homo sapiens (Human))
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Institute of Molecular Science and Technology (ISTM)
Curated by ChEMBL
Affinity DataKi: >300nMAssay Description:Binding affinity to human full length recombinant His-tagged HuR expressed in Escherichia coli Rosetta DH5alpha assessed as inhibition of interaction...More data for this Ligand-Target Pair


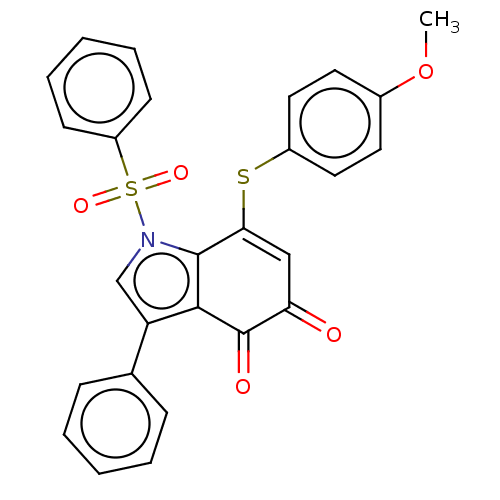
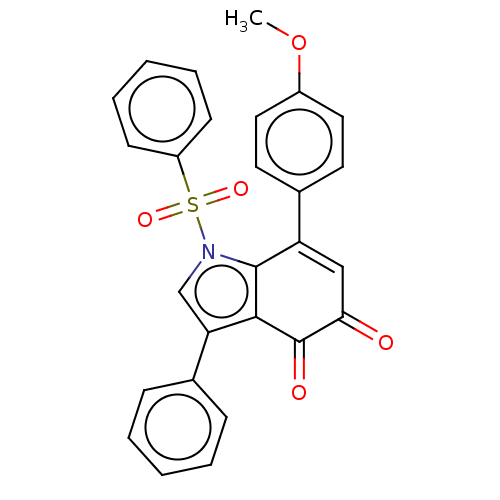



 3D Structure (crystal)
3D Structure (crystal)