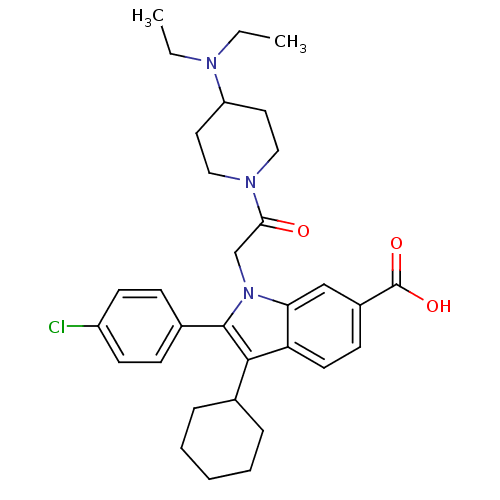
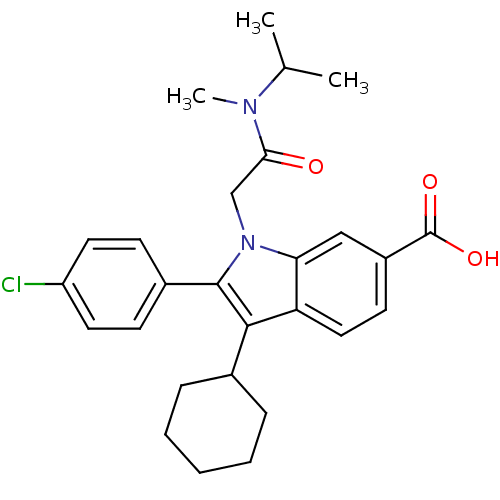
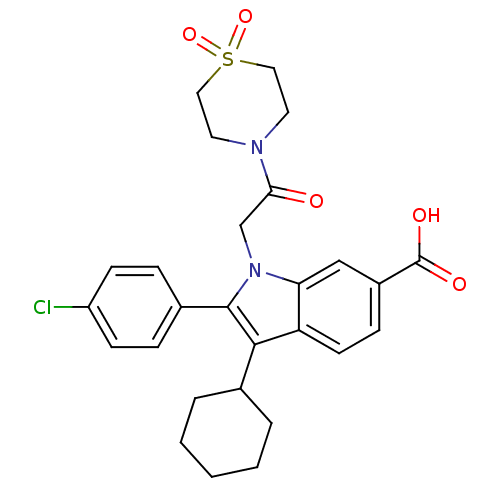
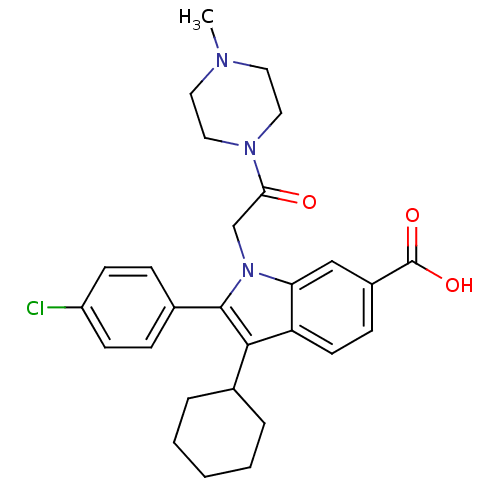
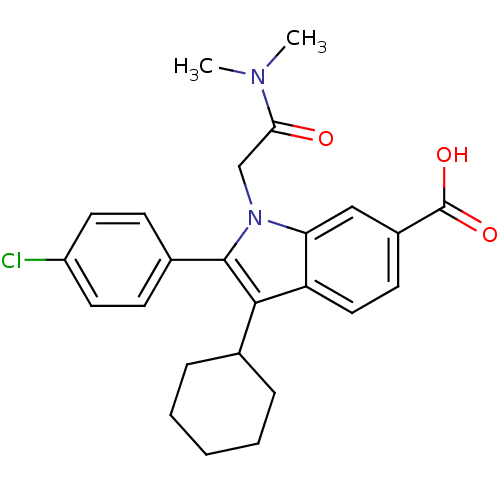
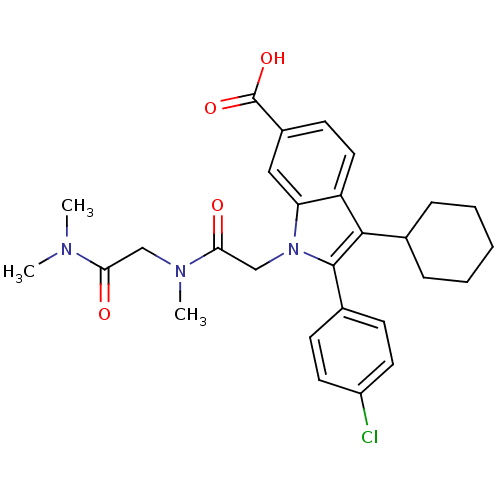
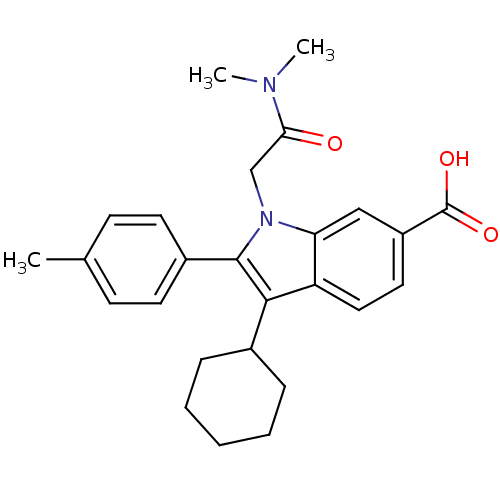
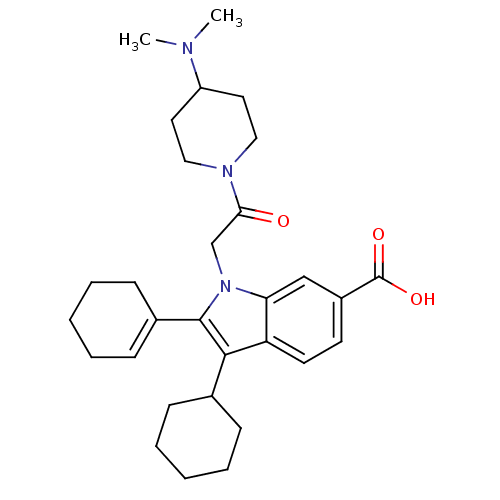
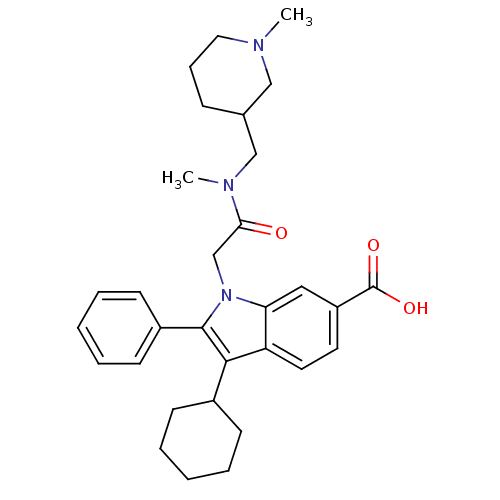
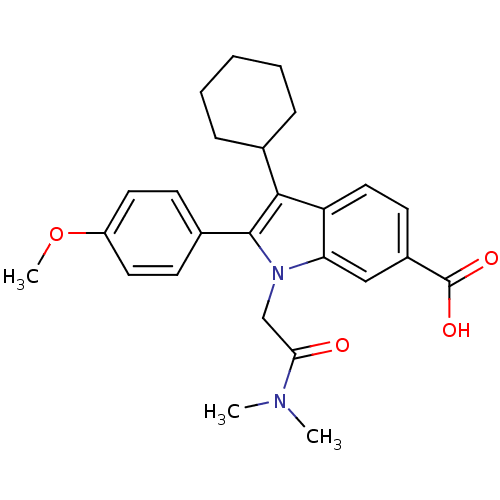
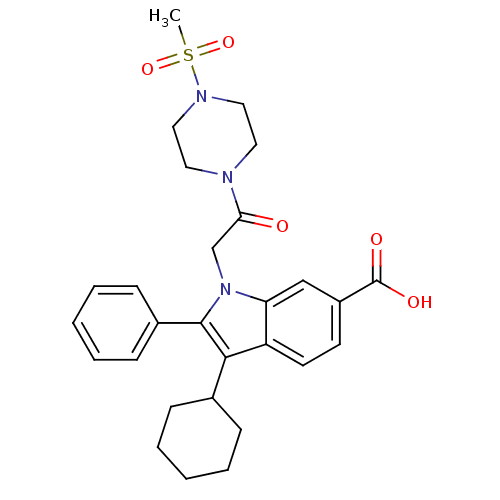
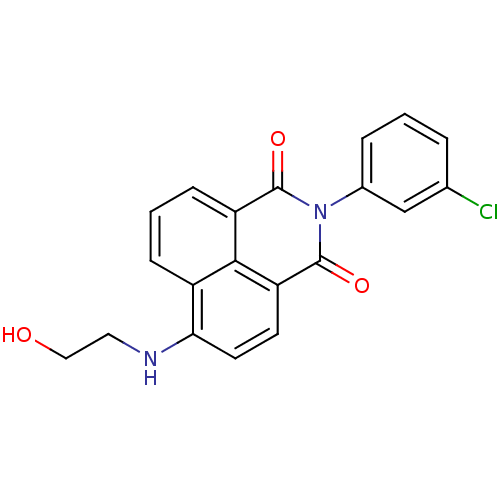
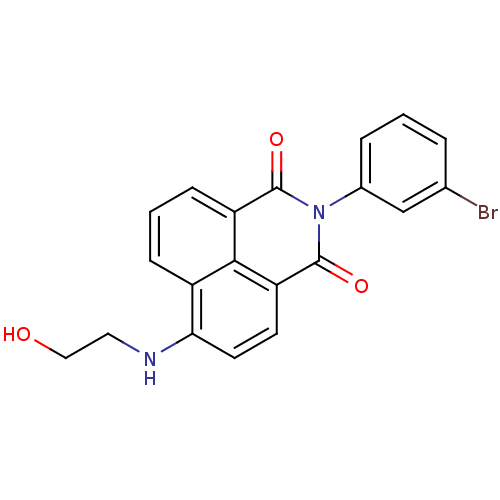
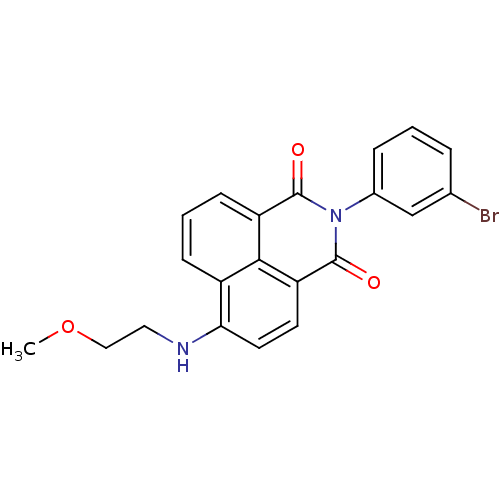
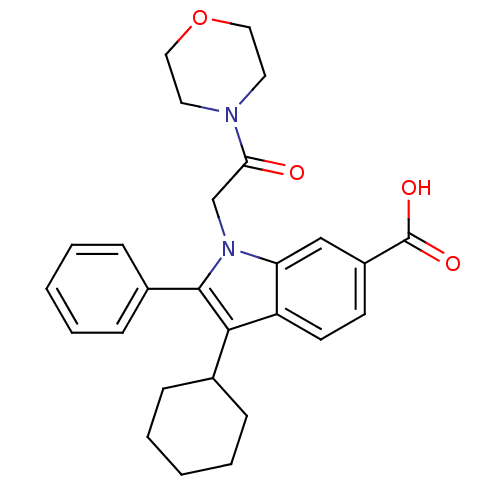
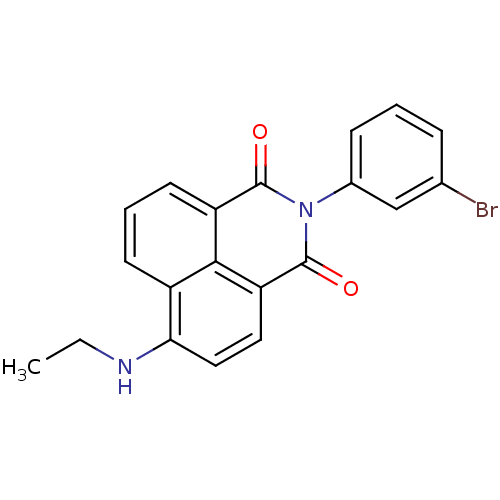
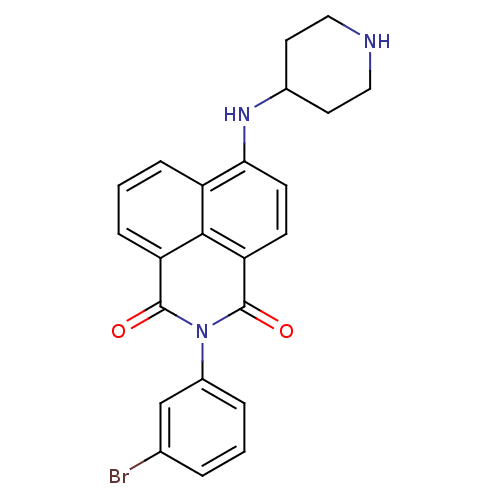
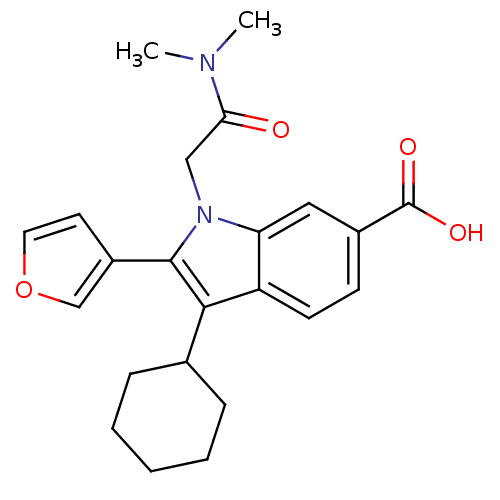
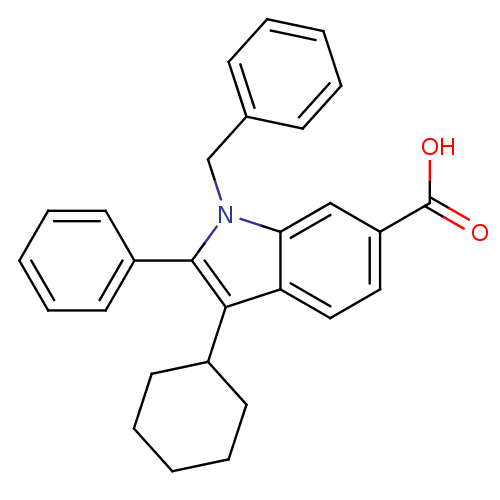
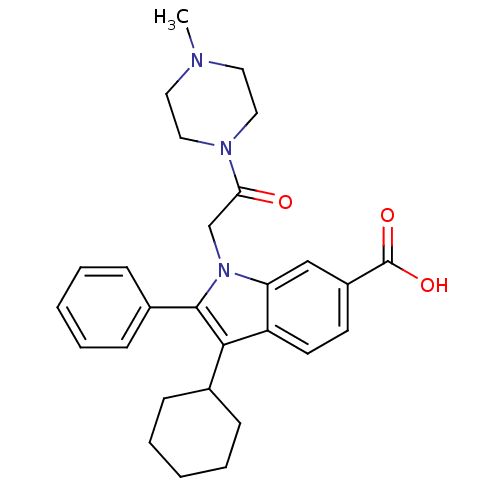
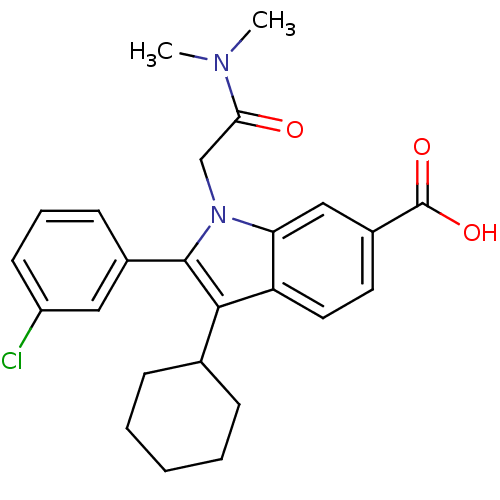
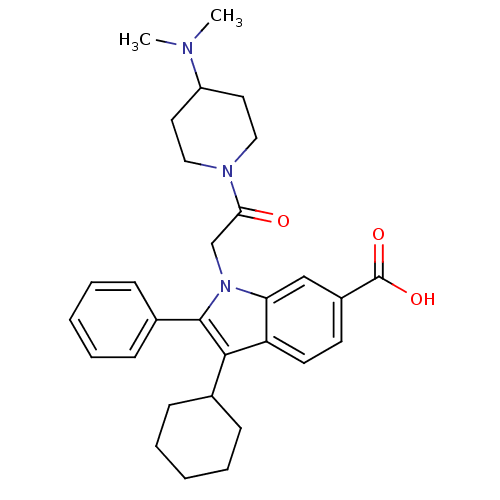
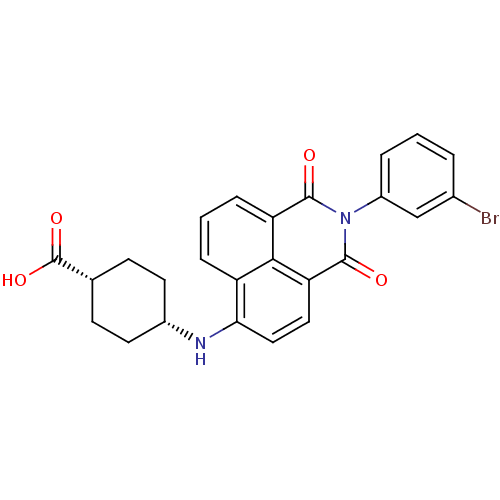
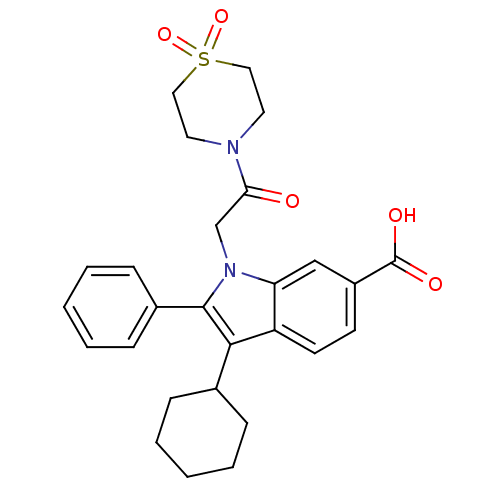
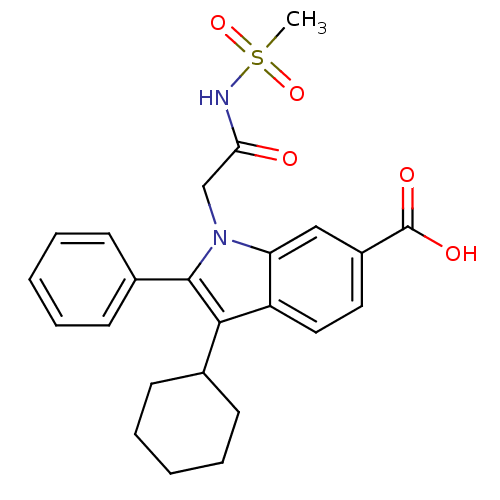
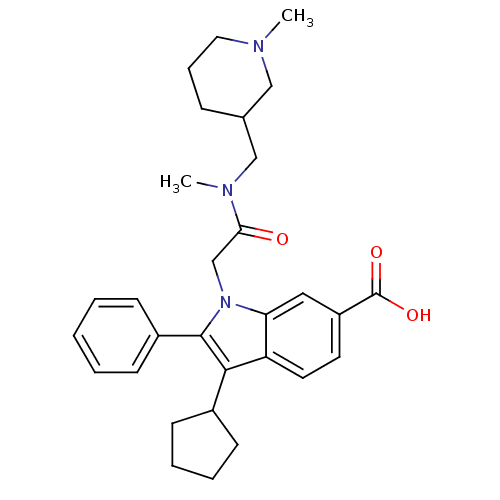
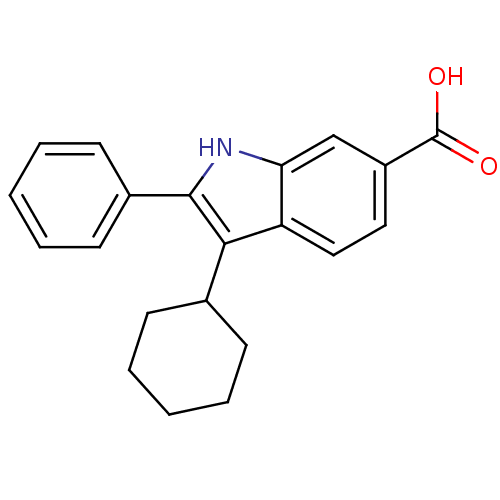
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 1.40nMAssay Description:Inhibitory concentration against RNA dependent RNA polymerase nonstructural protein 5B of hepatitis C virusMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 3nMAssay Description:Inhibitory concentration against RNA dependent RNA polymerase nonstructural protein 5B of hepatitis C virusMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 4.80nMAssay Description:Inhibitory concentration against RNA dependent RNA polymerase nonstructural protein 5B of hepatitis C virusMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 6nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 6nMAssay Description:Inhibitory concentration against RNA dependent RNA polymerase nonstructural protein 5B of hepatitis C virusMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 7nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 8nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 9nMAssay Description:Inhibitory activity against hepatitis C virus NS5B polymerase enzyme; Range=6-9 nMMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 9nMAssay Description:Inhibitory activity against hepatitis C virus NS5B polymerase enzyme; Range=6-9 nMMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 9nMAssay Description:Inhibitory activity against hepatitis C virus NS5B polymerase enzyme; Range=6-9 nMMore data for this Ligand-Target Pair
Affinity DataIC50: 9nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 9nMAssay Description:Inhibitory activity against hepatitis C virus NS5B polymerase enzyme; Range=6-9 nMMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 9nMAssay Description:Inhibitory activity against hepatitis C virus NS5B polymerase enzyme; Range=6-9 nMMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 11nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 11nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 11nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 12nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 12nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 13nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
Affinity DataIC50: 14nM EC50: 2.20E+3nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 15nMAssay Description:Inhibitory concentration against NS5B polymerase of Hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetGenome polyprotein [2420-2989,M2833T,R2963Q](Hepatitis C virus genotype 1b (isolate BK) (HCV))
IRBM-MRL Rome
IRBM-MRL Rome
Affinity DataIC50: 16nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 17nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 17nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 18nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 18nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 18nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 18nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 18nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 19nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
Affinity DataIC50: 19nM EC50: 3.80E+3nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetGenome polyprotein [2420-2989,P2914A,R2963Q](Hepatitis C virus genotype 1b (isolate BK) (HCV))
IRBM-MRL Rome
IRBM-MRL Rome
Affinity DataIC50: 20nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
Affinity DataIC50: 20nM EC50: 7.10E+3nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
Affinity DataIC50: 24nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 26nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 28nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
Affinity DataIC50: 29nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
Affinity DataIC50: 30nM EC50: 1.90E+3nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 31nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 37nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 39nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetGenome polyprotein [2420-2989,P2914A,R2963Q](Hepatitis C virus genotype 1b (isolate BK) (HCV))
IRBM-MRL Rome
IRBM-MRL Rome
Affinity DataIC50: 40nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 40nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 41nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
Affinity DataIC50: 42nM EC50: 3.80E+3nMpH: 7.5 T: 2°CAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 43nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
Affinity DataIC50: 43nMAssay Description:Primer-dependent assays were performed using the homopolymeric template/primer. Compounds, polymerase and template RNA were incubated at RT for 25 mi...More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 45nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 47nMAssay Description:Inhibitory activity against by hepatitis C virus NS5B polymeraseMore data for this Ligand-Target Pair
TargetRNA-directed RNA polymerase(Hepatitis C virus)
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
IRBM (Merck Research Laboratories Rome)
Curated by ChEMBL
Affinity DataIC50: 48nMAssay Description:Inhibitory activity against RNA dependent RNA polymerase nonstructural protein 5B in hepatitis C virus; n=1More data for this Ligand-Target Pair



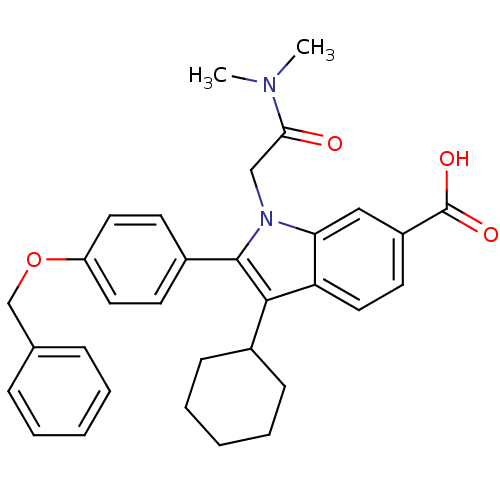

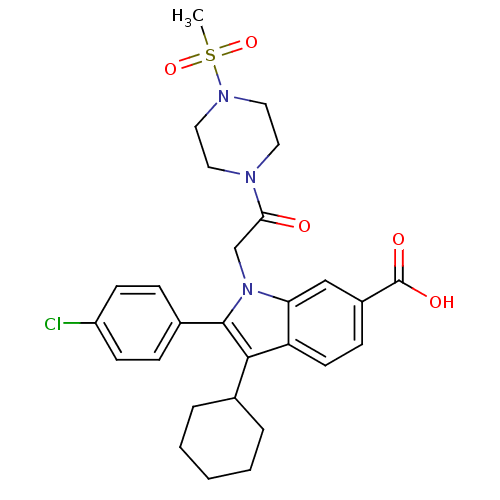



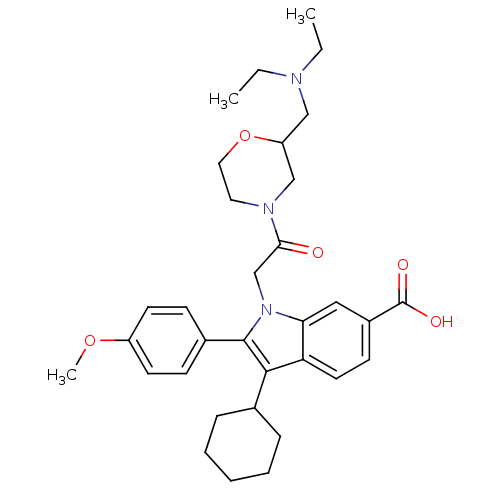

 3D Structure (crystal)
3D Structure (crystal)