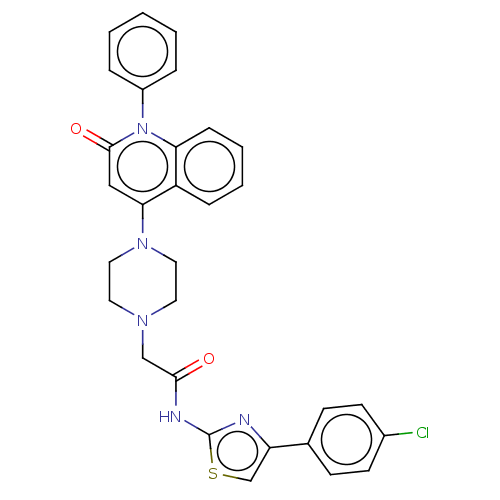
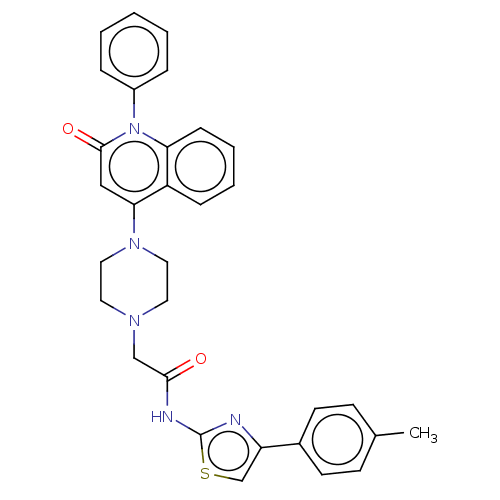
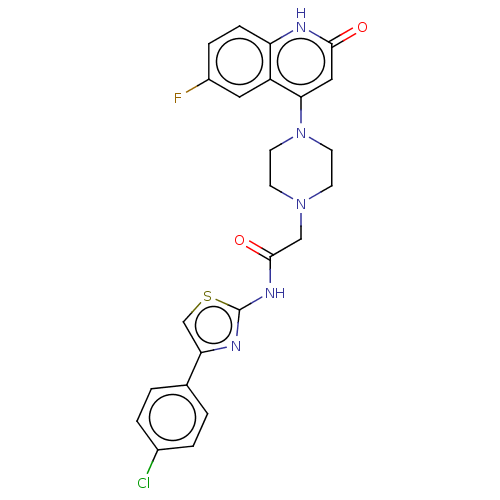
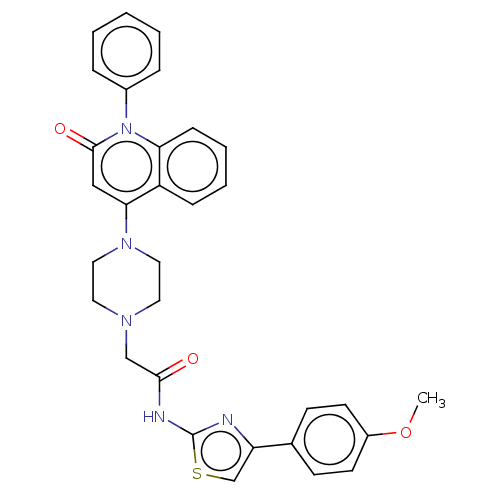
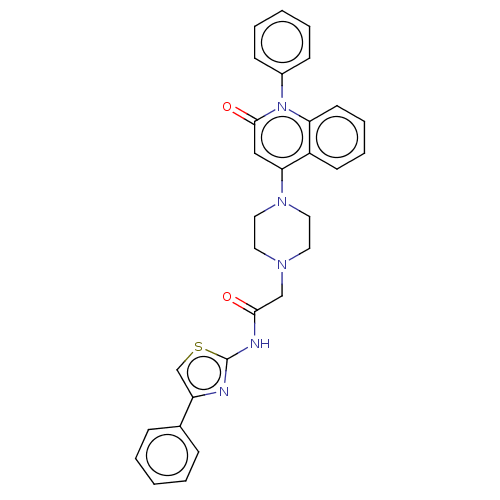
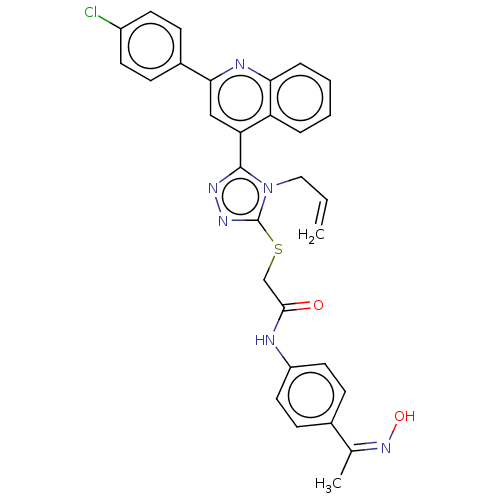
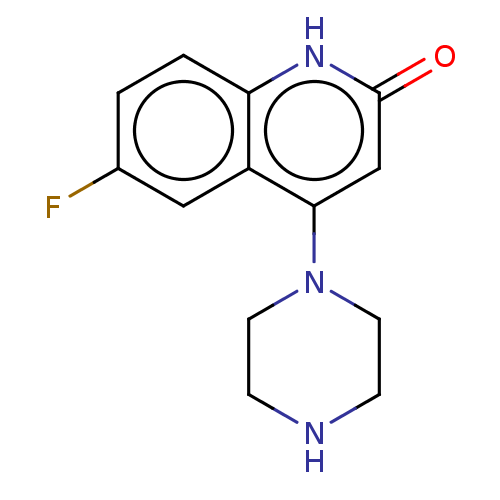
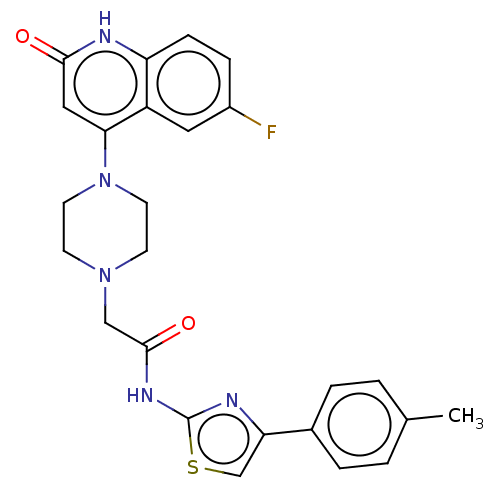
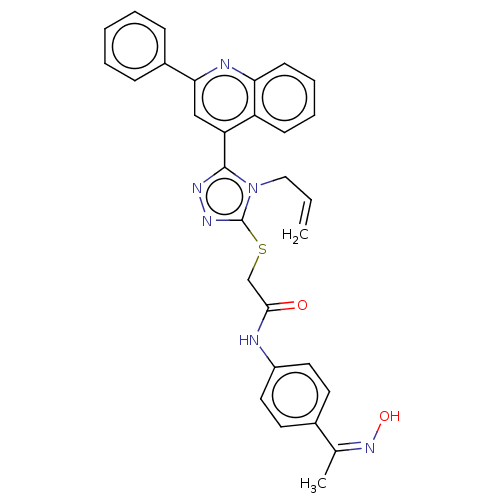
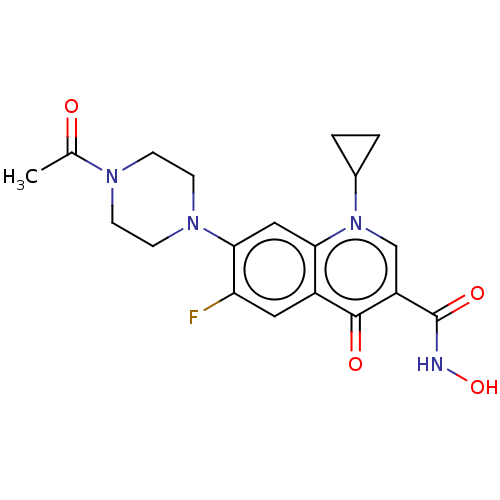
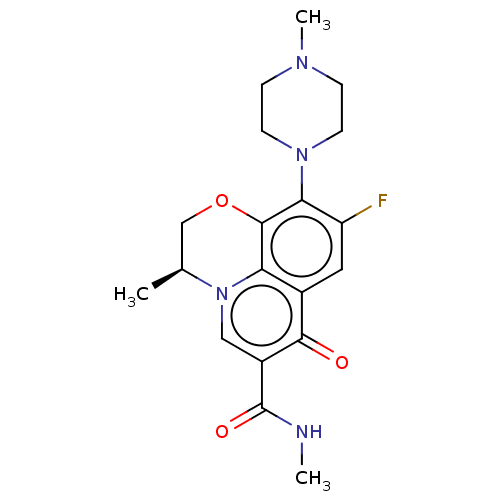
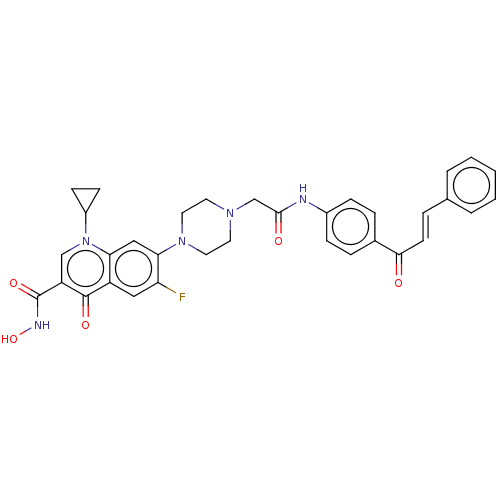
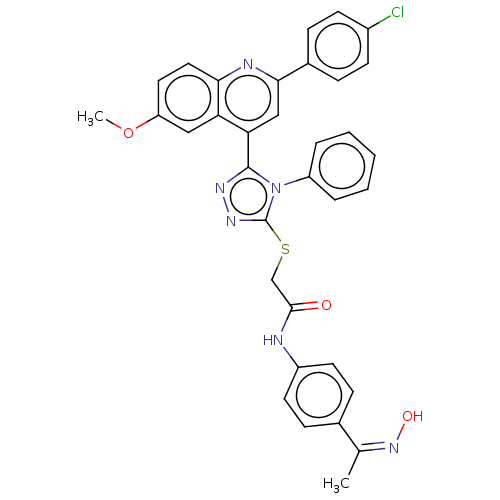
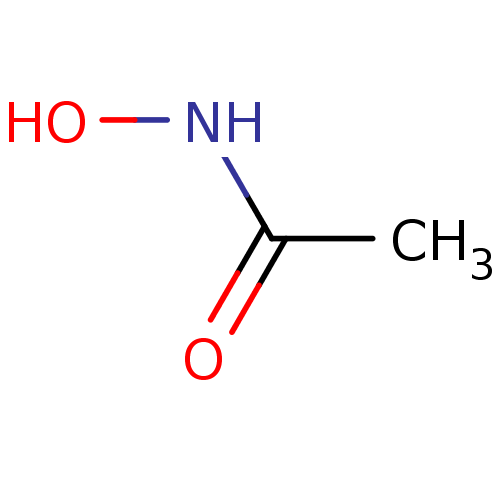
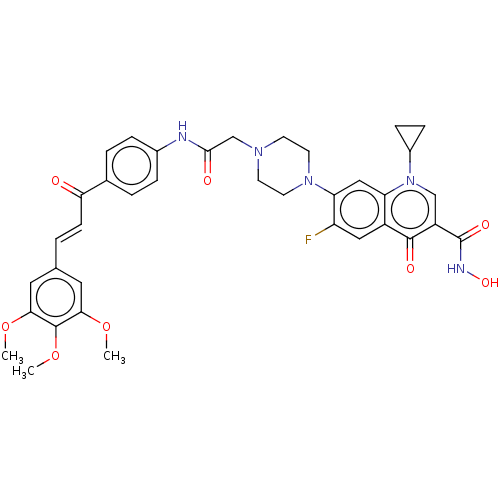
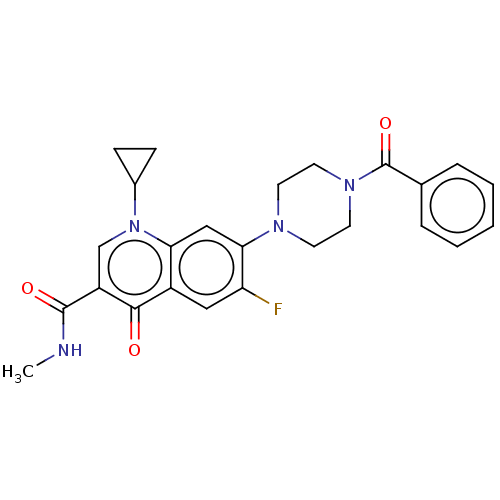
Affinity DataIC50: 47nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 51nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 51nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 63nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 112nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 167nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 170nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 175nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 223nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 250nMAssay Description:Displacement of FAM-labeled 5-GpYLPQTV-NH2 from recombinant GST-tagged STAT3 (unknown origin) preincubated for 30 mins followed by FAM-labeled 5-GpYL...More data for this Ligand-Target Pair
Affinity DataIC50: 282nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 296nMAssay Description:Inhibition of VEGFR-2 (unknown origin) by using poly (Glu:Tyr 4:1) as substrate measured after 45 mins by Kinase-Glo max based microplate readerMore data for this Ligand-Target Pair
Affinity DataIC50: 320nMAssay Description:Displacement of FAM-labeled 5-GpYLPQTV-NH2 from recombinant GST-tagged STAT3 (unknown origin) preincubated for 30 mins followed by FAM-labeled 5-GpYL...More data for this Ligand-Target Pair
Affinity DataIC50: 1.22E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.29E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.08E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.20E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.22E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.26E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.89E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.92E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.70E+3nMAssay Description:Inhibition of STAT3 in human SB-590885-resistant 451Lu cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-lu...More data for this Ligand-Target Pair
Affinity DataIC50: 6.30E+3nMAssay Description:Inhibition of STAT3 in human SB-590885-sensitive 451Lu cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-lu...More data for this Ligand-Target Pair
Affinity DataIC50: 7.17E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.90E+3nMAssay Description:Inhibition of STAT3 in human SB-590885-sensitive 451Lu cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-lu...More data for this Ligand-Target Pair
Affinity DataIC50: 9.90E+3nMAssay Description:Inhibition of STAT3 in human SB-590885-resistant MEL1617 cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-...More data for this Ligand-Target Pair
Affinity DataIC50: 1.03E+4nMAssay Description:Inhibition of STAT3 in human SB-590885-sensitive MEL1617 cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-...More data for this Ligand-Target Pair
Affinity DataIC50: 1.56E+4nMAssay Description:Inhibition of STAT3 in human SB-590885-resistant MEL1617 cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-...More data for this Ligand-Target Pair
Affinity DataIC50: 1.70E+4nMAssay Description:Inhibition of STAT3 in human SB-590885-resistant 451Lu cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-lu...More data for this Ligand-Target Pair
Affinity DataIC50: 1.89E+4nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.93E+4nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.14E+4nMAssay Description:Inhibition of STAT3 in human SB-590885-sensitive MEL1617 cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-...More data for this Ligand-Target Pair
Affinity DataIC50: 3.52E+4nMAssay Description:Inhibition of STAT3 in human SB-590885-resistant MEL1617 cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-...More data for this Ligand-Target Pair
Affinity DataIC50: 4.05E+4nMAssay Description:Inhibition of STAT3 in human SB-590885-resistant 451Lu cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-lu...More data for this Ligand-Target Pair
Affinity DataIC50: 5.40E+4nMAssay Description:Inhibition of STAT3 in human SB-590885-sensitive 451Lu cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-lu...More data for this Ligand-Target Pair
Affinity DataIC50: 7.50E+4nMAssay Description:Inhibition of STAT3 in human SB-590885-sensitive MEL1617 cells assessed as reduction in STAT3 transcriptional activity measured after 12 hrs by Dual-...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+5nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.31E+5nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.72E+5nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 8.12E+5nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 9.12E+5nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 9.49E+5nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.41E+6nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+6nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)