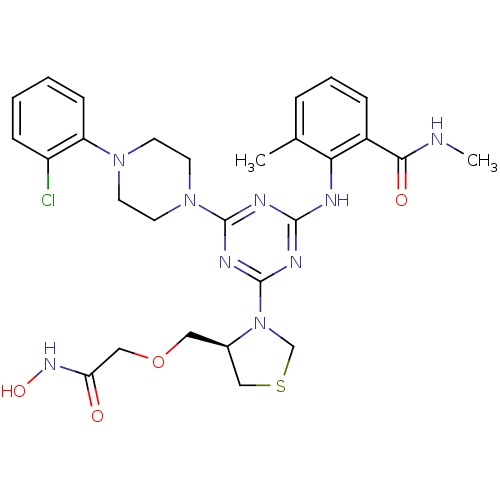
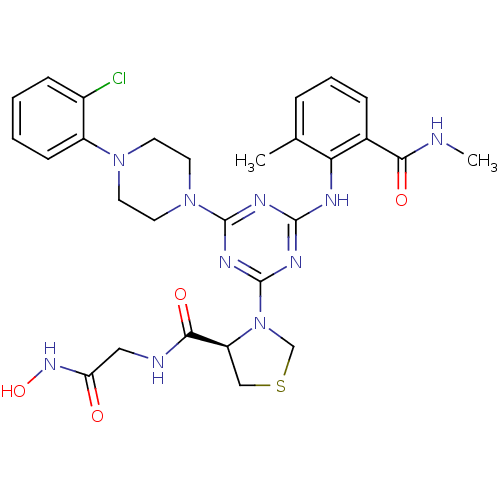
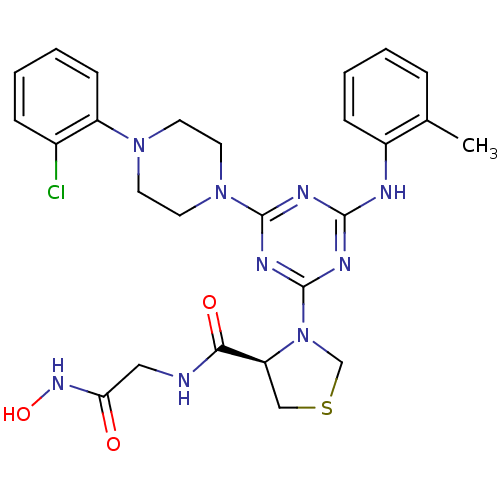
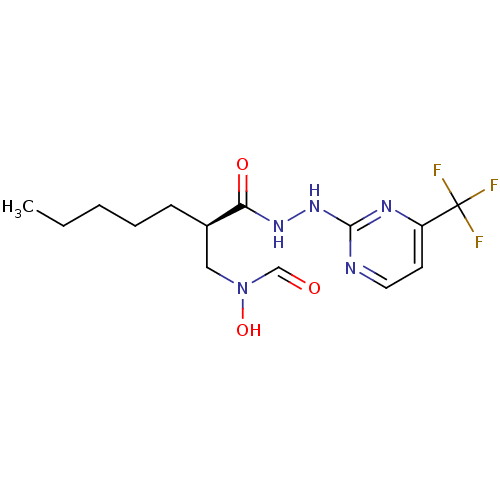
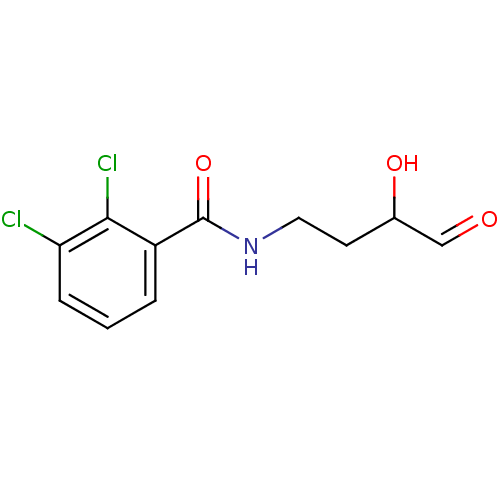
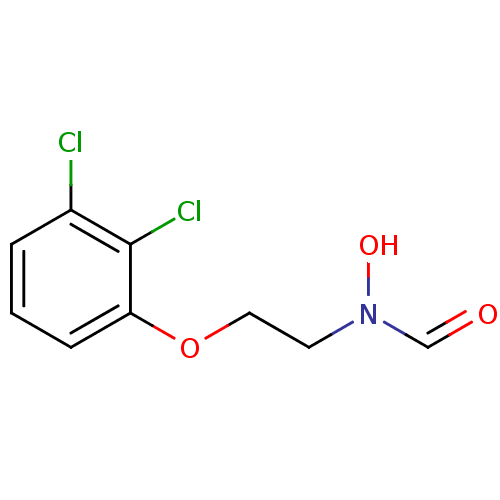
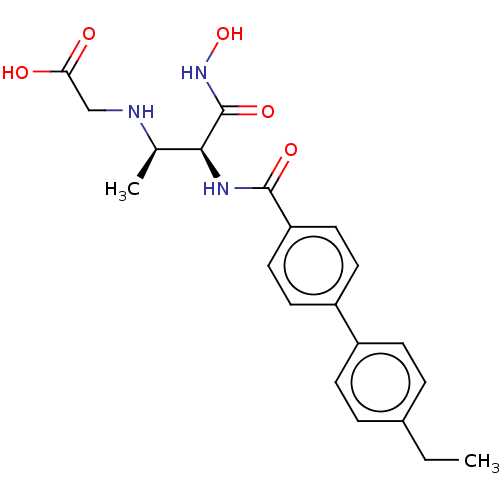
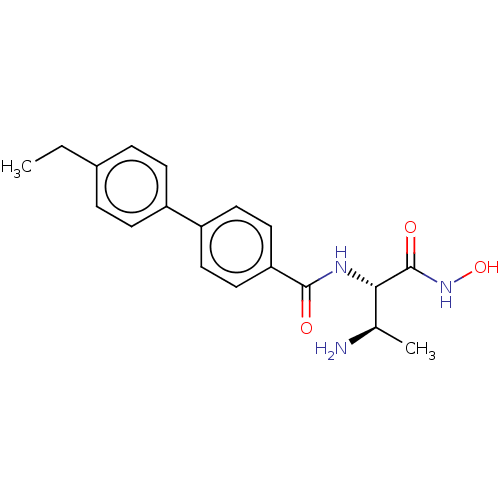
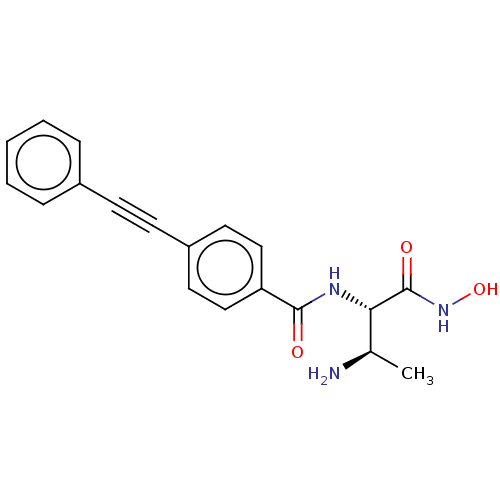
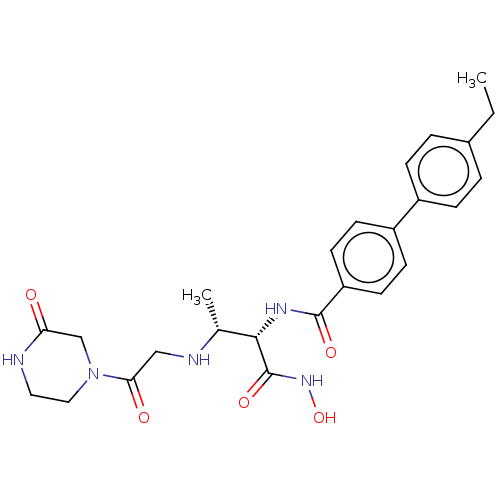
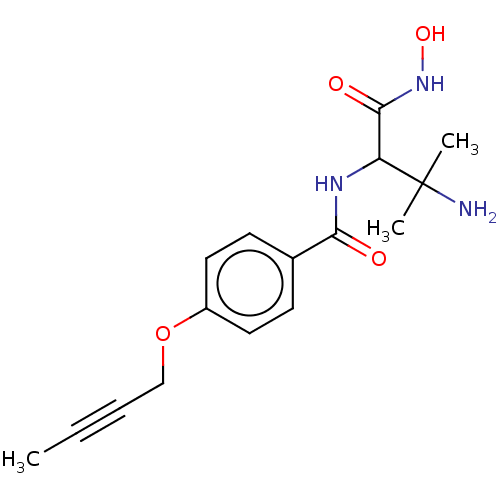
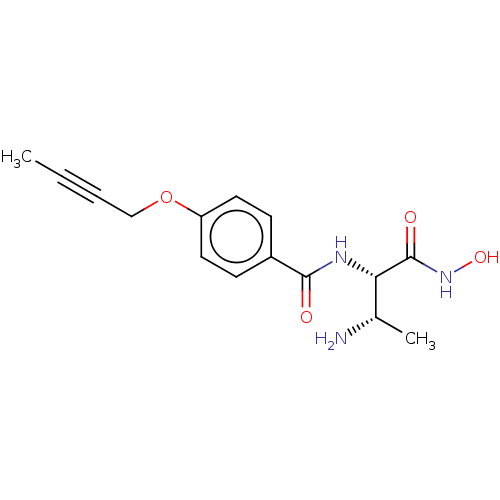
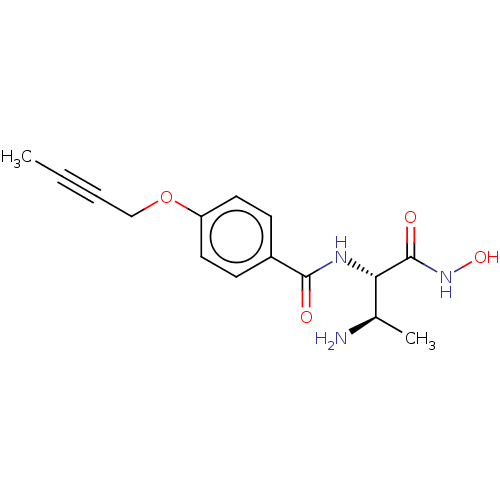
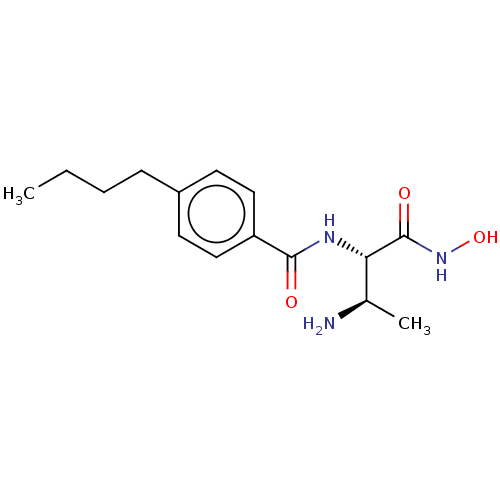
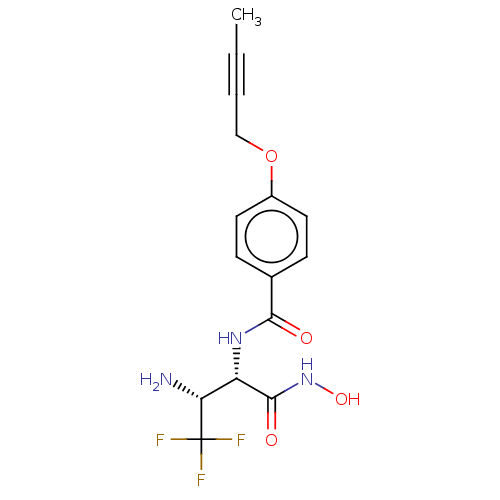
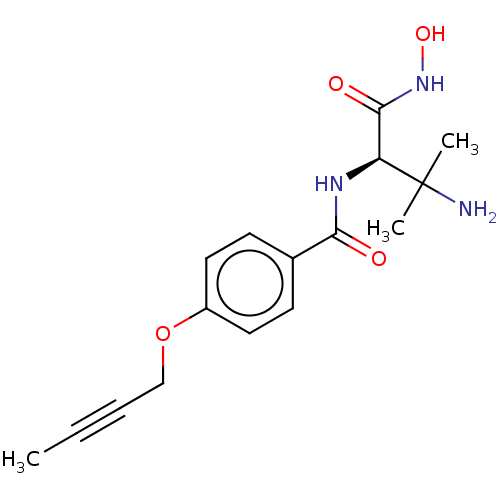
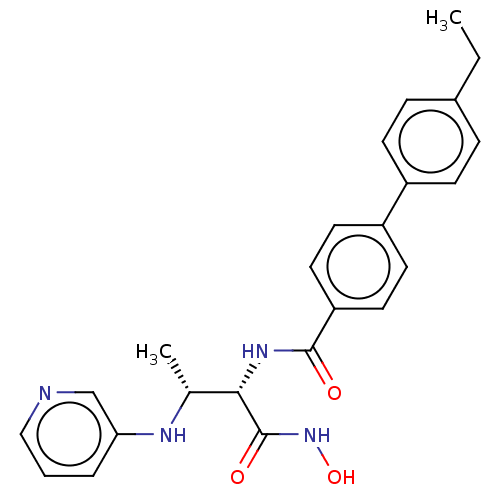
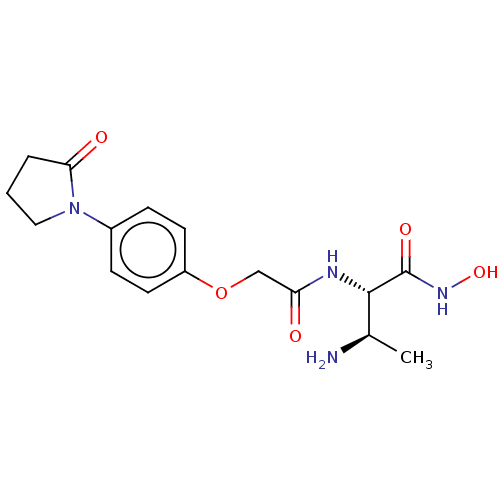
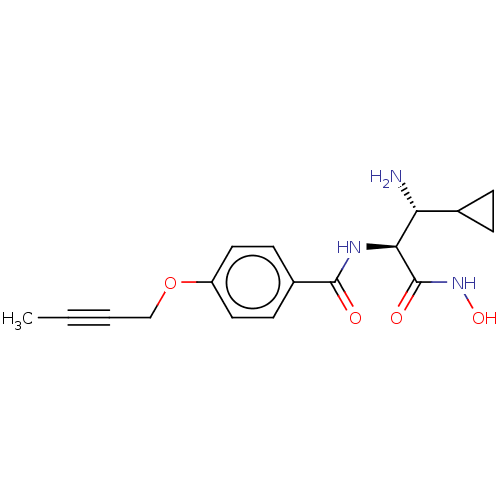
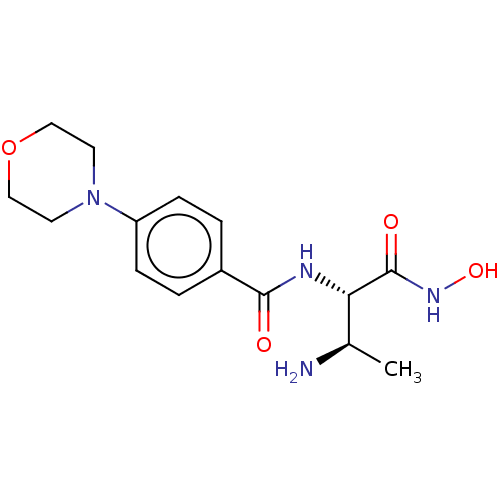
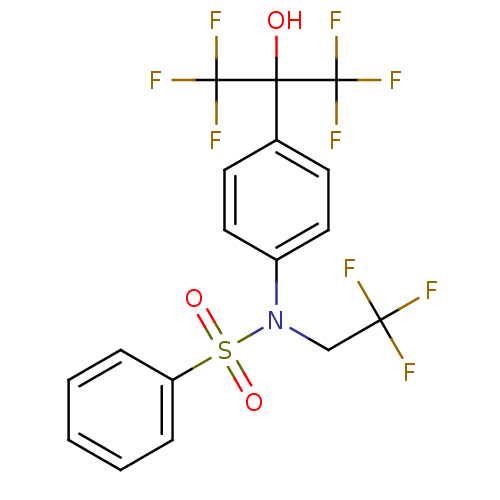
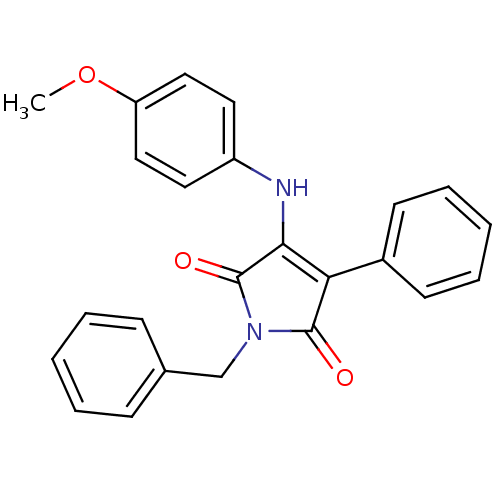
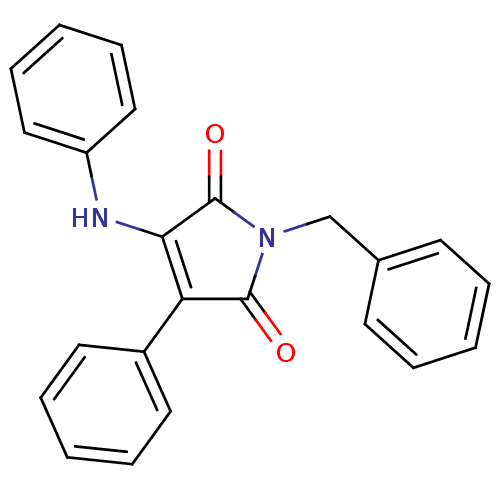
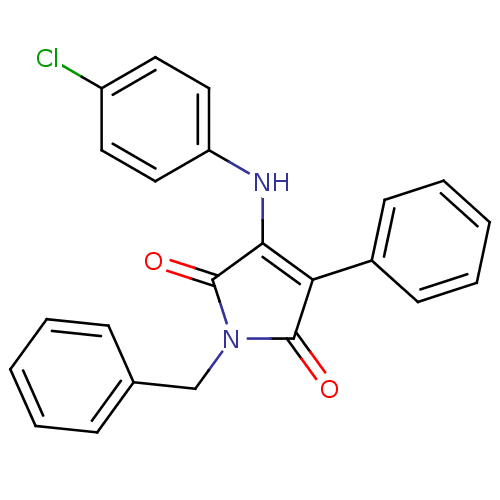
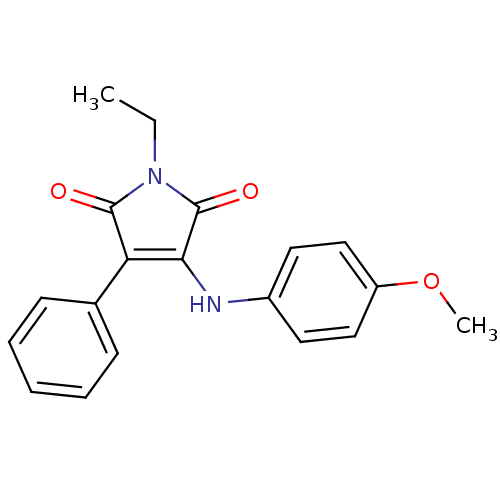
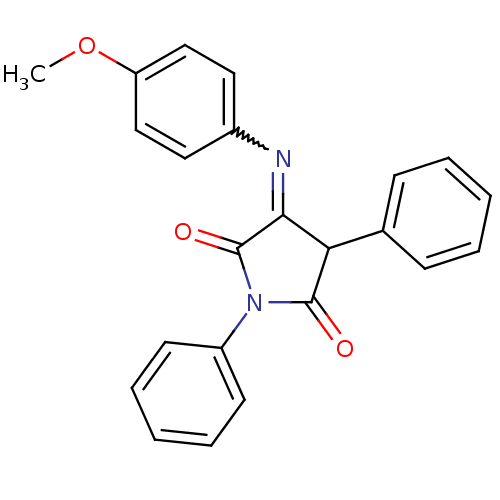
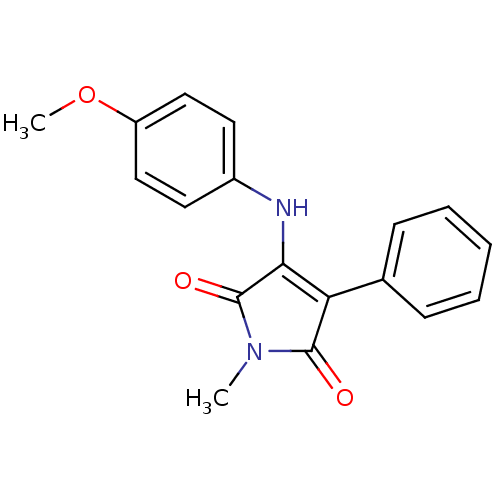
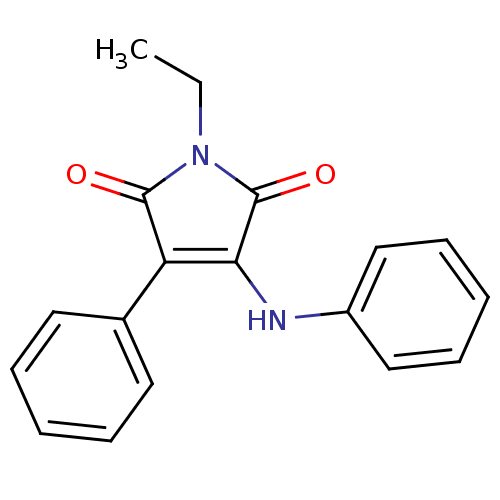
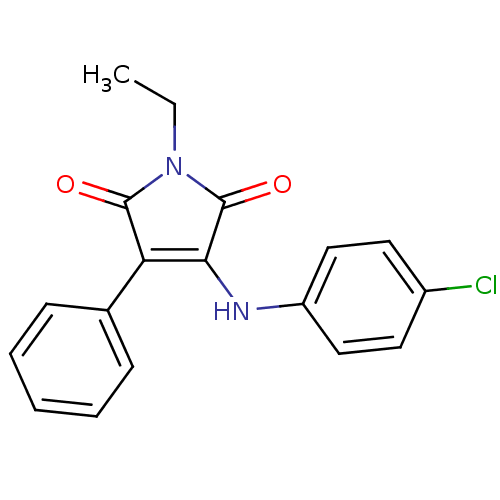
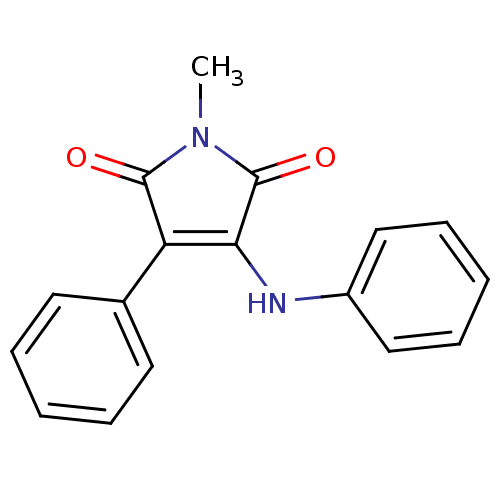
Affinity DataKi: 53.7nMAssay Description:Binding assay using PDF inhibitors.More data for this Ligand-Target Pair
Affinity DataKi: 117nMAssay Description:Binding assay using PDF inhibitors.More data for this Ligand-Target Pair
Affinity DataKi: 213nMAssay Description:Binding assay using PDF inhibitors.More data for this Ligand-Target Pair
Affinity DataKi: 334nMAssay Description:Binding assay using PDF inhibitors.More data for this Ligand-Target Pair
Affinity DataKi: 650nMAssay Description:Binding assay using PDF inhibitors.More data for this Ligand-Target Pair
Affinity DataKi: 6.31E+3nMAssay Description:Binding assay using PDF inhibitors.More data for this Ligand-Target Pair
Affinity DataKi: 2.59E+4nMAssay Description:Binding assay using PDF inhibitors.More data for this Ligand-Target Pair
Affinity DataKi: 3.17E+4nMAssay Description:Binding assay using PDF inhibitors.More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 1nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 1.46nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.13nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.56nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 4.15nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 4.19nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 12.5nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 16.4nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 19.7nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 37.5nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 39nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 81.4nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 98nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 116nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 268nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 342nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 362nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 899nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa)
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 1.64E+3nMAssay Description:Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 30 mins followed by sub...More data for this Ligand-Target Pair
TargetNuclear factor erythroid 2-related factor 2(Homo sapiens (Human))
The State University of New Jersey
Curated by ChEMBL
The State University of New Jersey
Curated by ChEMBL
Affinity DataIC50: 3.00E+3nMAssay Description:Inhibition of Keap1-Nrf2 interaction (unknown origin) after 30 mins by fluorescence polarization assayMore data for this Ligand-Target Pair
TargetHistone deacetylase 1(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC1More data for this Ligand-Target Pair
TargetHistone deacetylase 2(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC2More data for this Ligand-Target Pair
TargetHistone deacetylase 4(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC4More data for this Ligand-Target Pair
TargetHistone deacetylase 5(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC5More data for this Ligand-Target Pair
TargetPolyamine deacetylase HDAC10(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC10More data for this Ligand-Target Pair
TargetHistone deacetylase 9(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC9More data for this Ligand-Target Pair
TargetHistone deacetylase 7(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC7More data for this Ligand-Target Pair
TargetHistone deacetylase 6(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC6More data for this Ligand-Target Pair
TargetHistone deacetylase 3(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC3More data for this Ligand-Target Pair
TargetHistone deacetylase 8(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC8More data for this Ligand-Target Pair
TargetHistone deacetylase 11(Homo sapiens (Human))
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Novartis Institutes for BioMedical Research
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human HDAC11More data for this Ligand-Target Pair
Affinity DataEC50: 80nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 55nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 50nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 100nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 130nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 300nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 275nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 575nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 890nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 725nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair
Affinity DataEC50: 1.59E+3nMpH: 7.5 T: 2°CAssay Description:The LXR LiSA measures the ligand-dependent recruitment of a 25 amino acid fragment of the steroid receptor coactivator 1 (SRC1) to the ligand-binding...More data for this Ligand-Target Pair

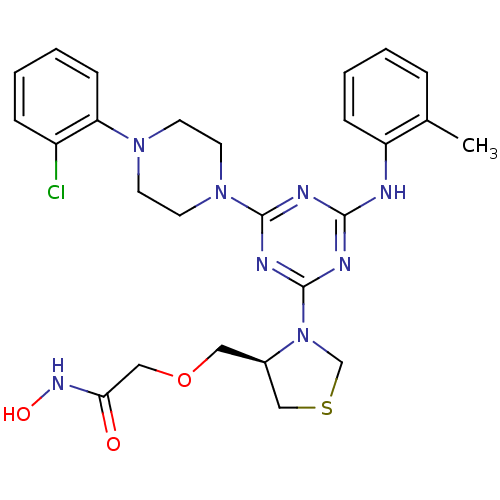
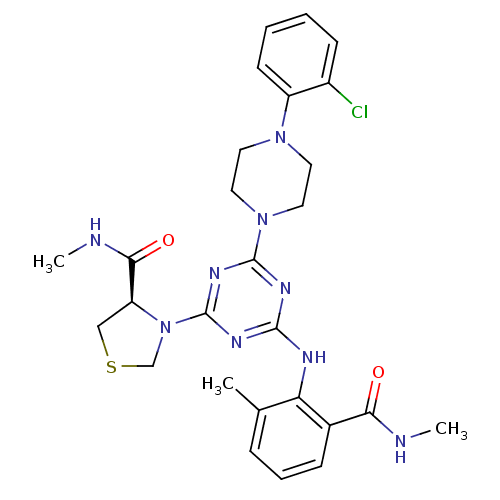
 3D Structure (crystal)
3D Structure (crystal)