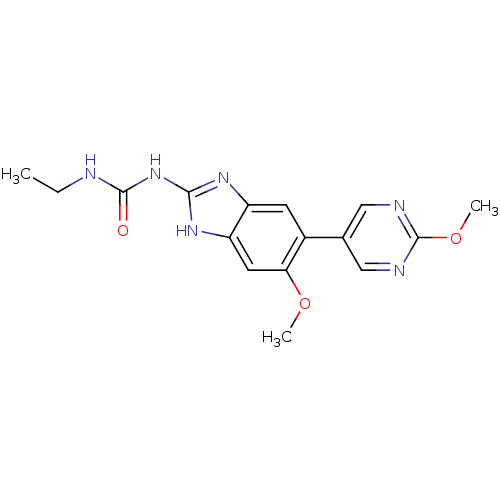
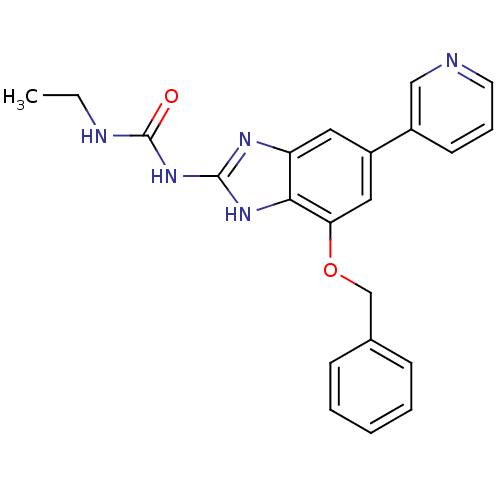
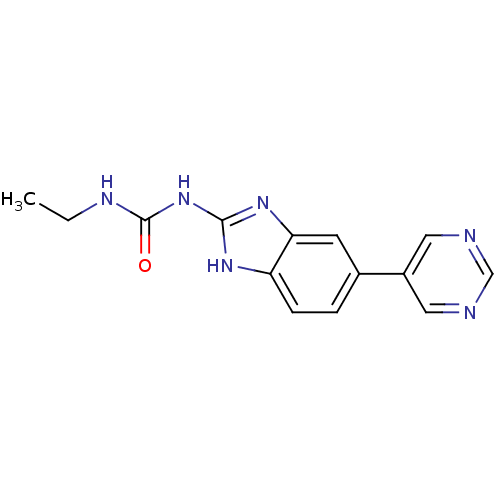
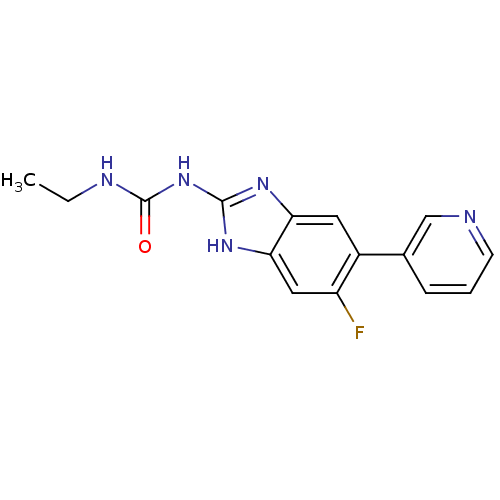
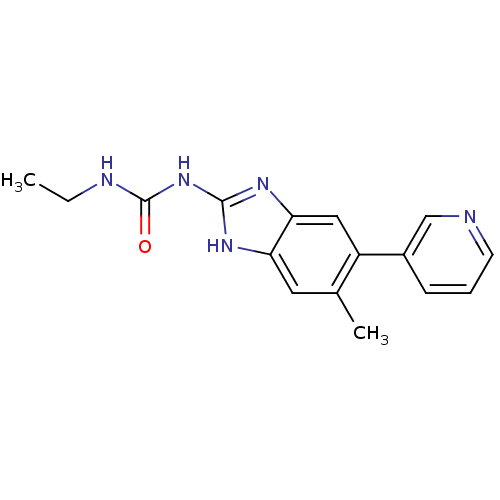
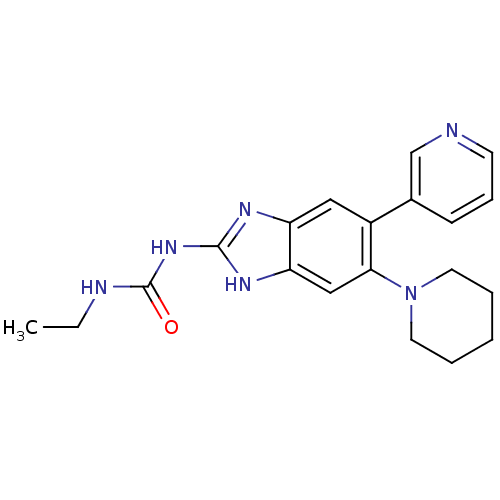
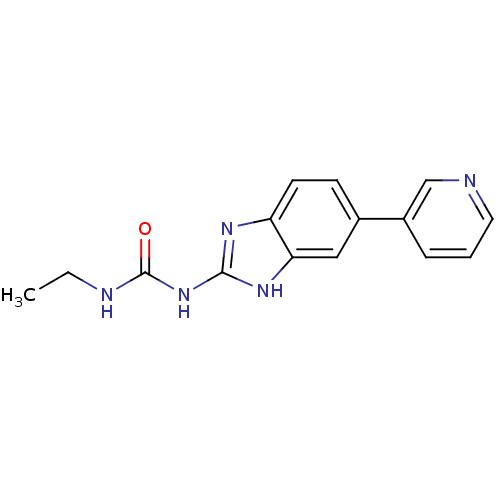
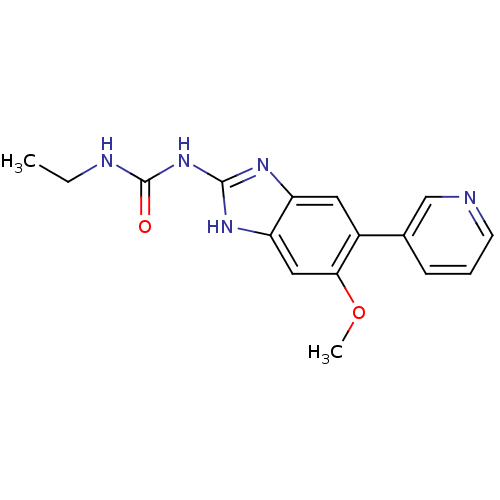
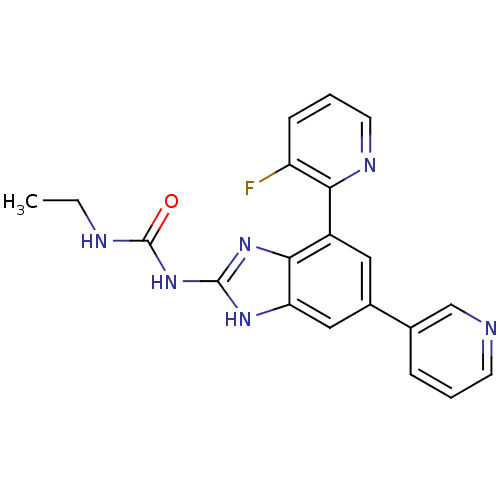
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
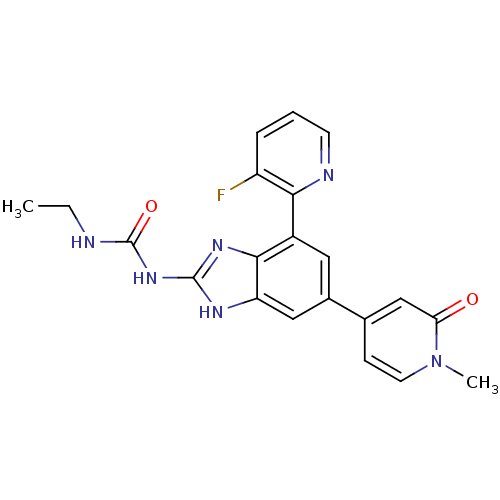
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
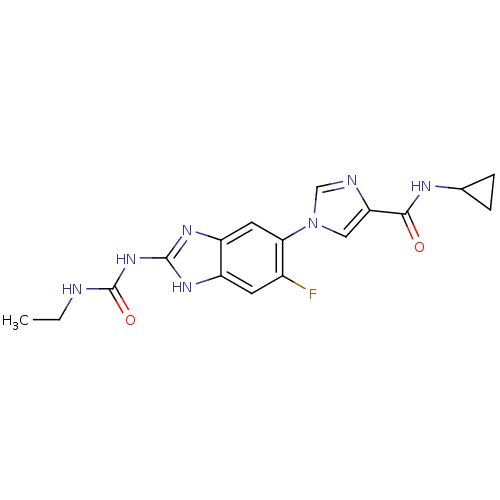
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
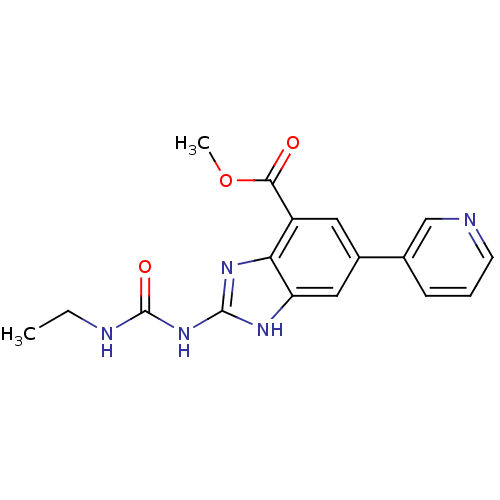
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <4nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 5nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 6nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 7nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 7nM ΔG°: -47.3kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 8nM ΔG°: -47.0kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nM ΔG°: -46.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nM ΔG°: -46.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nM ΔG°: -46.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 12nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 12nM ΔG°: -46.0kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 14nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 14nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 14nM ΔG°: -45.6kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 15nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 15nM ΔG°: -45.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 16nM ΔG°: -45.2kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 17nM ΔG°: -45.1kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 18nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 20nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 23nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 23nM ΔG°: -44.3kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 26nM ΔG°: -44.0kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 33nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 35nM ΔG°: -43.3kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 35nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 38nM ΔG°: -43.1kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 44nM ΔG°: -42.7kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 46nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 49nM ΔG°: -42.4kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 58nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 61nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 78nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 81nM ΔG°: -41.2kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 81nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 85nM ΔG°: -41.0kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 110nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 120nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 130nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 130nM ΔG°: -40.0kJ/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 150nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair

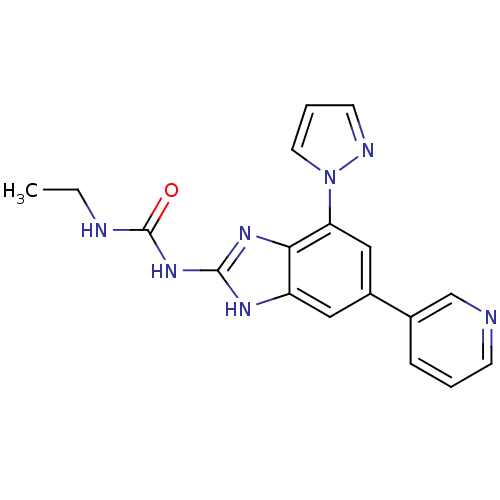
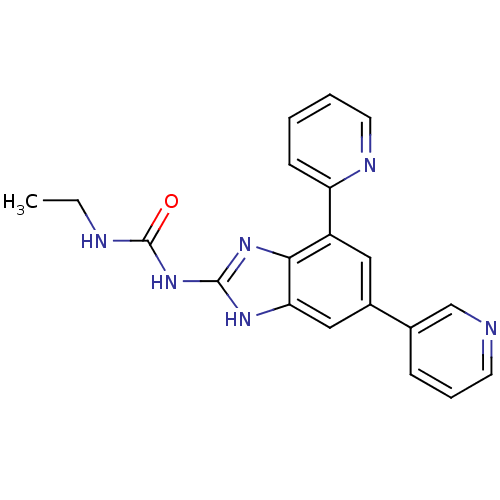
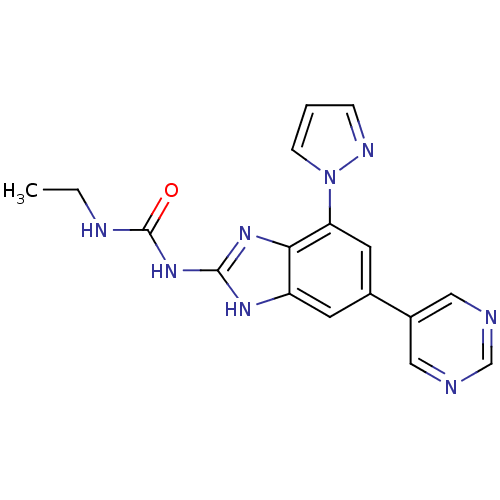
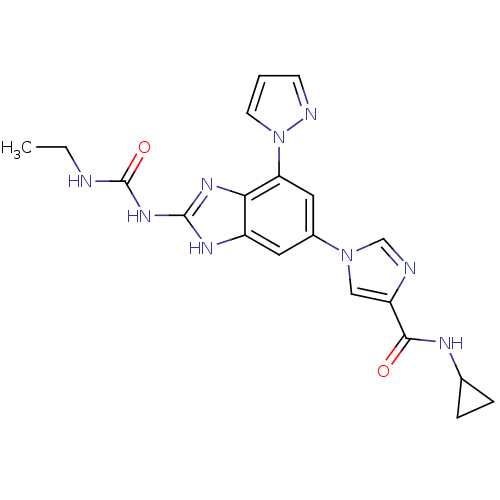
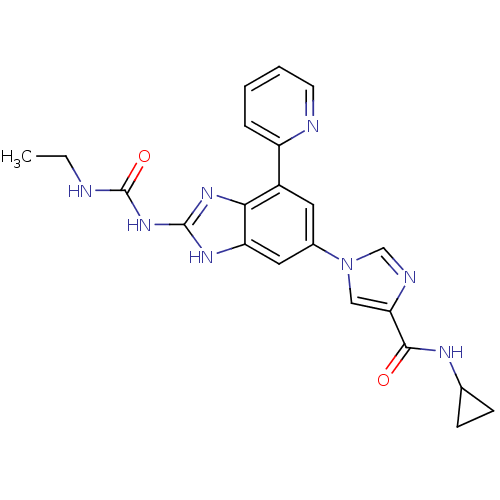
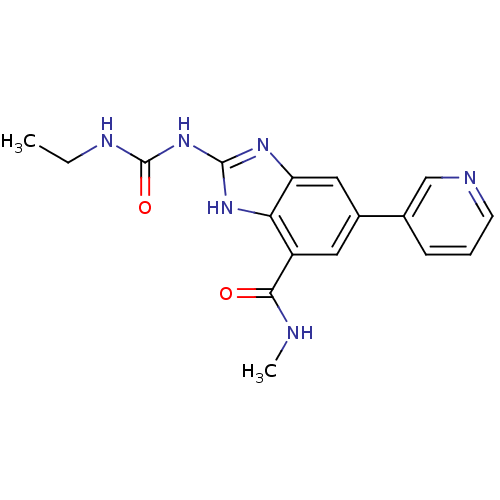
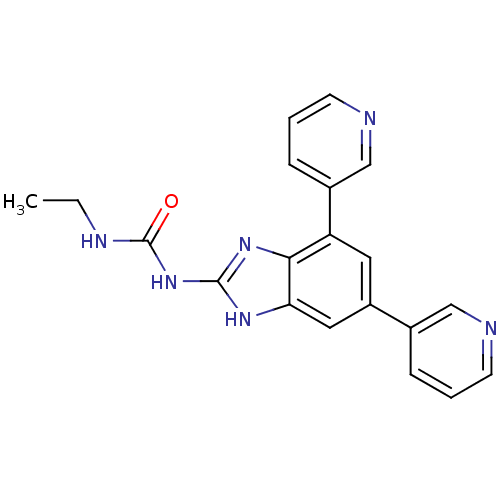
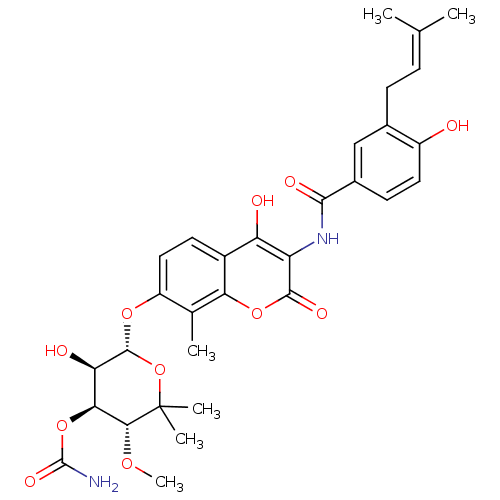
 3D Structure (crystal)
3D Structure (crystal)