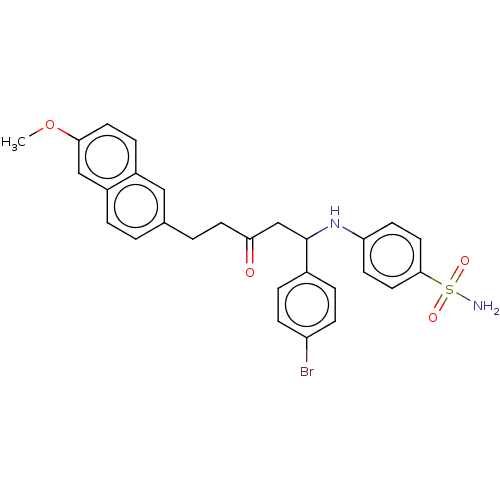
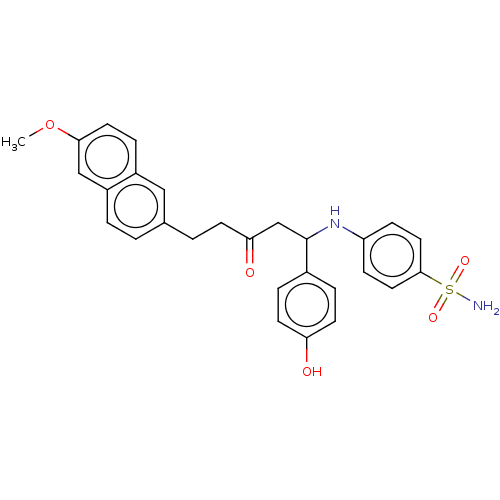
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
Chinese Academy of Sciences
Curated by ChEMBL
Chinese Academy of Sciences
Curated by ChEMBL
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
Northwest University
Curated by ChEMBL
Northwest University
Curated by ChEMBL
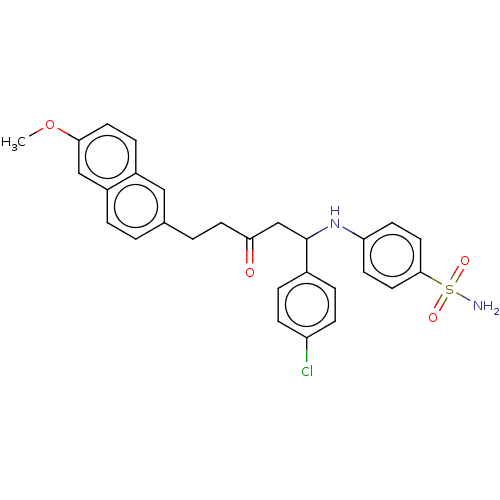
Affinity DataIC50: 560nMAssay Description:Inhibition of PTP1B (unknown origin) expressed in Escherichia coli expression system using p-nitrophenyl phosphate as substrate assessed as release o...More data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
Northwest University
Curated by ChEMBL
Northwest University
Curated by ChEMBL
Affinity DataIC50: 1.91E+3nMAssay Description:Inhibition of PTP1B (unknown origin) expressed in Escherichia coli expression system using p-nitrophenyl phosphate as substrate assessed as release o...More data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
Northwest University
Curated by ChEMBL
Northwest University
Curated by ChEMBL
Affinity DataIC50: 3.06E+3nMAssay Description:Inhibition of human recombinant PTP1B after 10 minsMore data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
Chinese Academy of Sciences
Curated by ChEMBL
Chinese Academy of Sciences
Curated by ChEMBL
Affinity DataIC50: 7.71E+3nMAssay Description:Inhibition of XODMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
Northwest University
Curated by ChEMBL
Northwest University
Curated by ChEMBL
Affinity DataIC50: 8.54E+3nMAssay Description:Inhibition of PTP1B (unknown origin) expressed in Escherichia coli expression system using p-nitrophenyl phosphate as substrate assessed as release o...More data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
Northwest University
Curated by ChEMBL
Northwest University
Curated by ChEMBL
Affinity DataIC50: 9.21E+3nMAssay Description:Inhibition of PTP1B (unknown origin) expressed in Escherichia coli expression system using p-nitrophenyl phosphate as substrate assessed as release o...More data for this Ligand-Target Pair
TargetXanthine dehydrogenase/oxidase(Homo sapiens (Human))
Chinese Academy of Sciences
Curated by ChEMBL
Chinese Academy of Sciences
Curated by ChEMBL
Affinity DataIC50: 1.15E+4nMAssay Description:Inhibition of XODMore data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
Northwest University
Curated by ChEMBL
Northwest University
Curated by ChEMBL
Affinity DataIC50: 1.22E+4nMAssay Description:Inhibition of PTP1B (unknown origin) expressed in Escherichia coli expression system using p-nitrophenyl phosphate as substrate assessed as release o...More data for this Ligand-Target Pair
TargetTyrosine-protein phosphatase non-receptor type 1(Homo sapiens (Human))
Northwest University
Curated by ChEMBL
Northwest University
Curated by ChEMBL
Affinity DataIC50: 1.61E+4nMAssay Description:Inhibition of PTP1B (unknown origin) expressed in Escherichia coli expression system using p-nitrophenyl phosphate as substrate assessed as release o...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha/delta/gamma(Homo sapiens (Human))
Southwest University
Curated by ChEMBL
Southwest University
Curated by ChEMBL
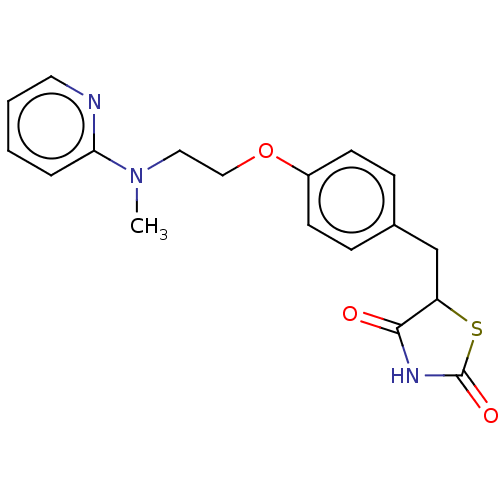
Affinity DataEC50: 1.28E+4nMAssay Description:Activation of PPAR in human HepG2 cells after 24 hrs by luciferase reporter gene assayMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha/delta/gamma(Homo sapiens (Human))
Southwest University
Curated by ChEMBL
Southwest University
Curated by ChEMBL
Affinity DataEC50: 120nMAssay Description:Activation of PPAR in human HepG2 cells after 24 hrs by luciferase reporter gene assayMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha/delta/gamma(Homo sapiens (Human))
Southwest University
Curated by ChEMBL
Southwest University
Curated by ChEMBL
Affinity DataEC50: 1.44E+4nMAssay Description:Activation of PPAR in human HepG2 cells after 24 hrs by luciferase reporter gene assayMore data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor alpha/delta/gamma(Homo sapiens (Human))
Southwest University
Curated by ChEMBL
Southwest University
Curated by ChEMBL
Affinity DataEC50: 1.10E+4nMAssay Description:Activation of PPAR in human HepG2 cells after 24 hrs by luciferase reporter gene assayMore data for this Ligand-Target Pair

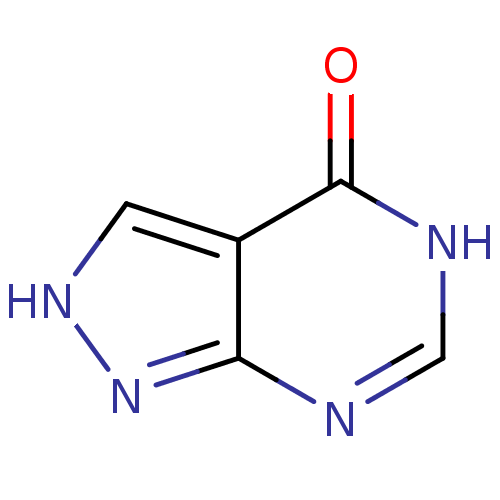
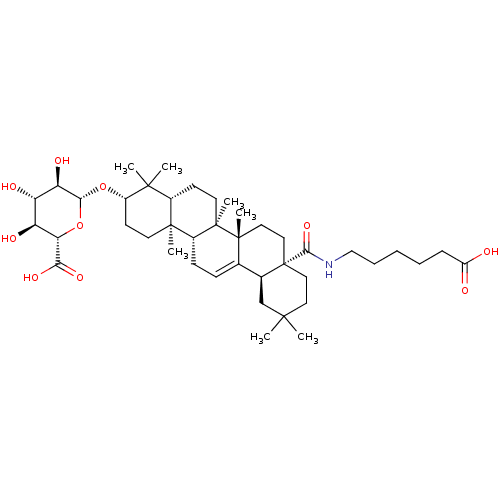
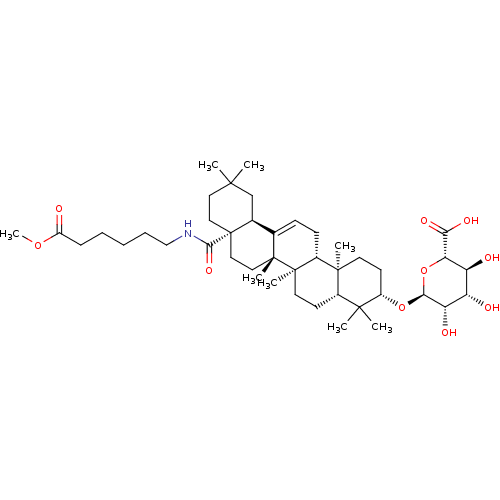
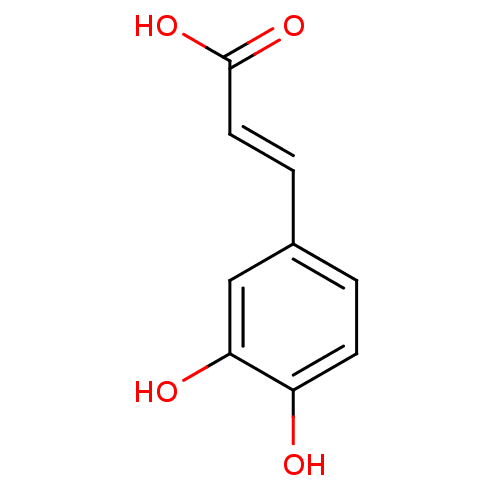
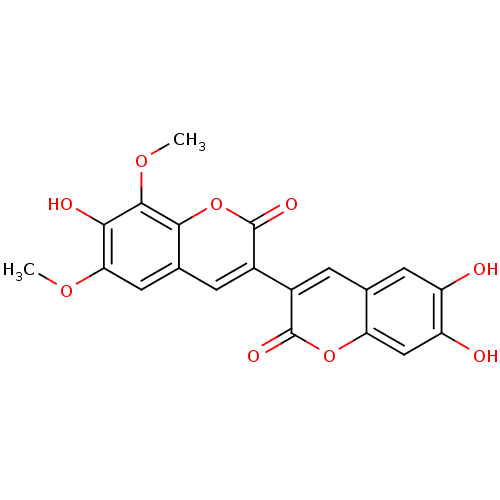
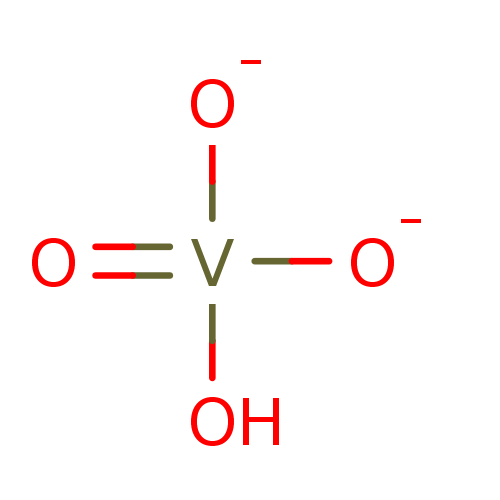
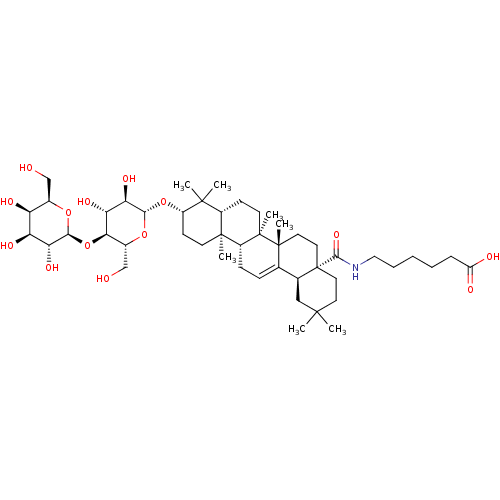
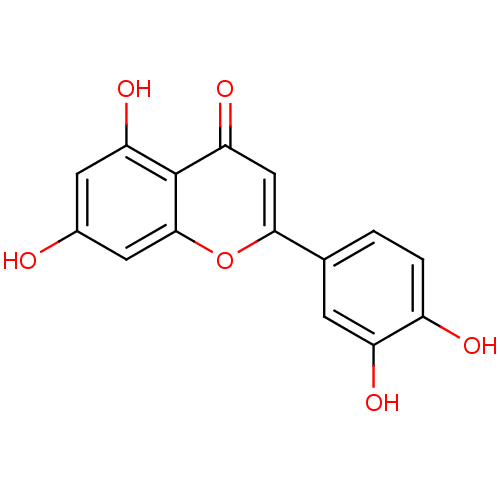
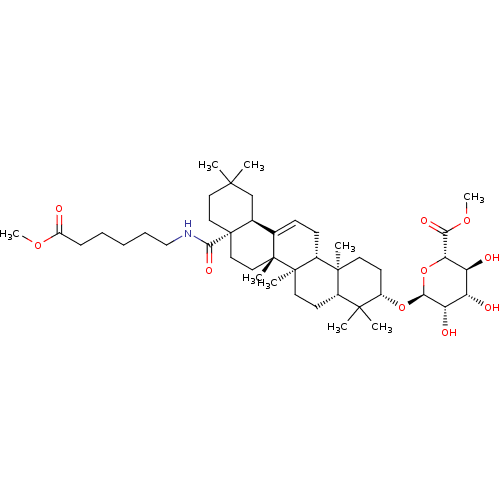
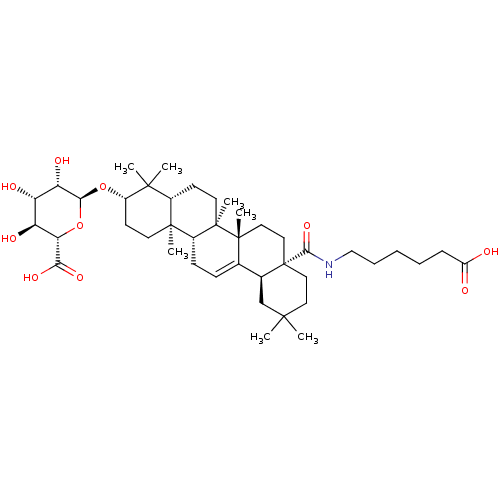
 3D Structure (crystal)
3D Structure (crystal)