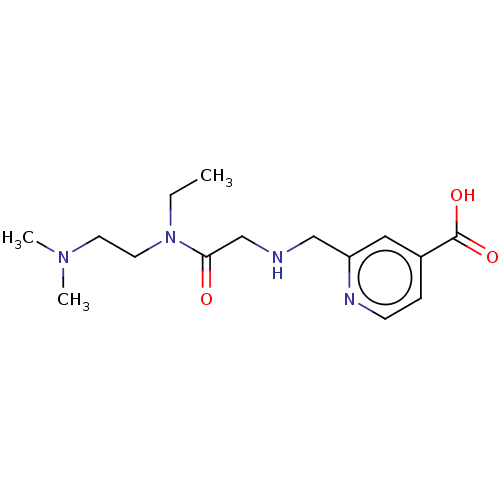
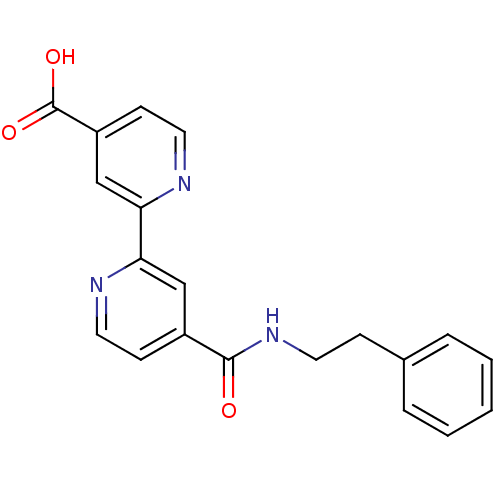
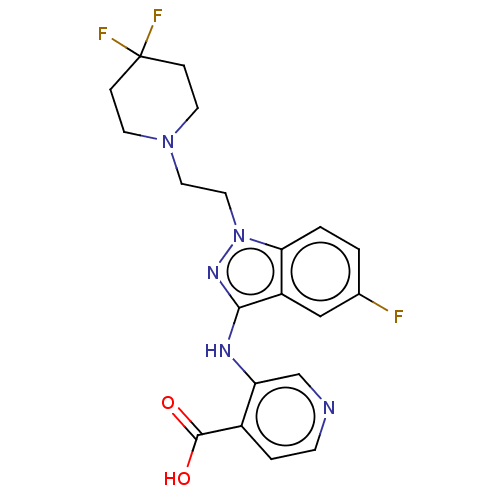
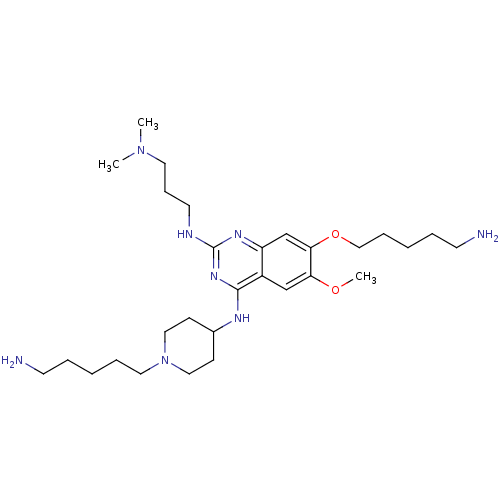
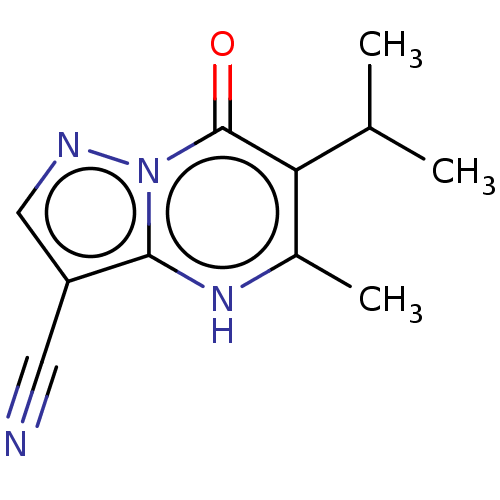
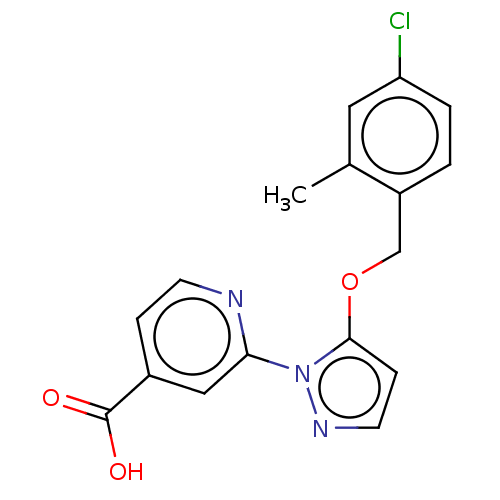
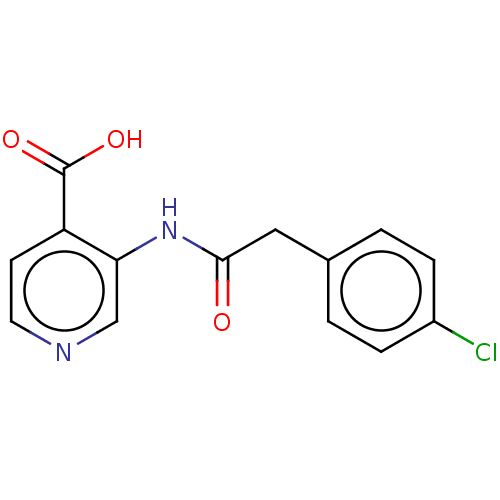
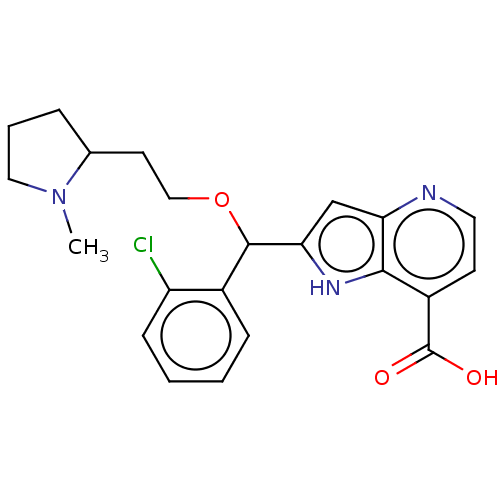
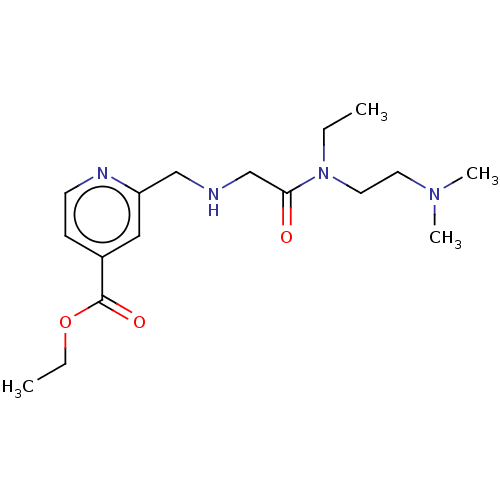
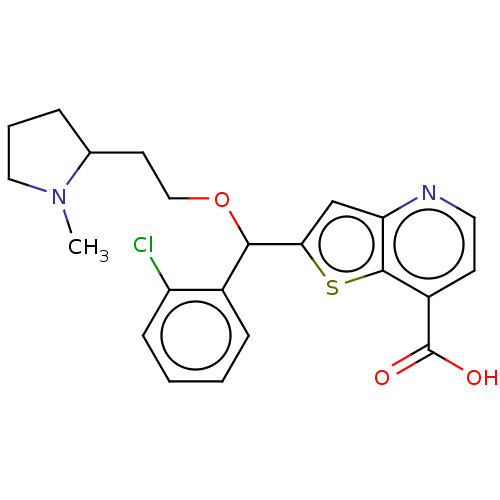
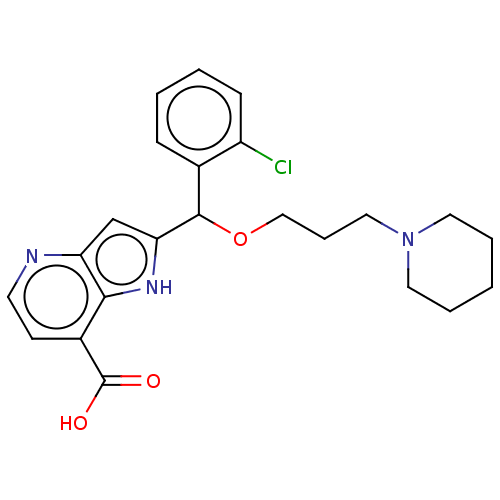
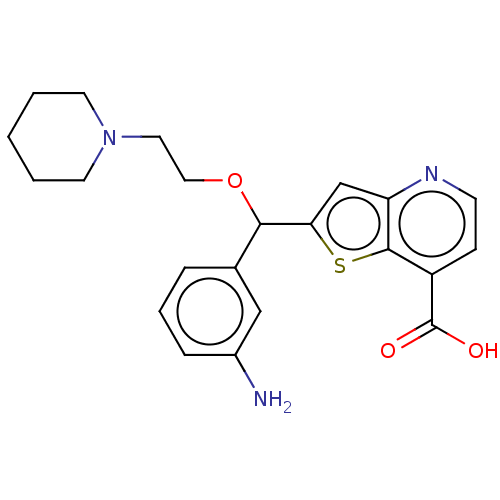
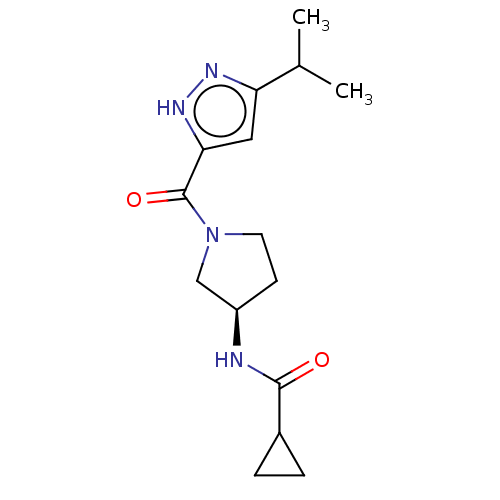
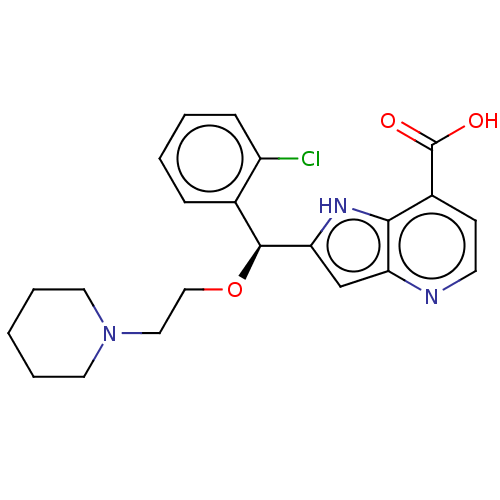
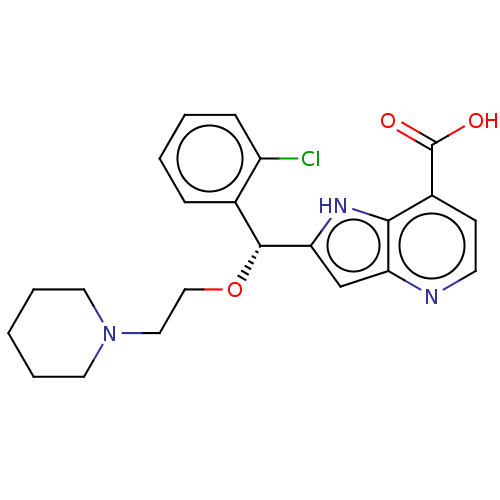
TargetHistone-lysine N-methyltransferase EHMT2(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKi: 3.60nMAssay Description:Competitive inhibition of recombinant human G9a using biotinylated-Histone H3 peptide (1 to 21 residues) as substrate after 30 mins in presence of va...More data for this Ligand-Target Pair
TargetHistone-lysine N-methyltransferase EHMT2(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKi: 14nMAssay Description:Non-competitive inhibition of recombinant human G9a using biotinylated-Histone H3 peptide (1 to 21 residues) as substrate at varying concentration af...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKi: 410nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin)More data for this Ligand-Target Pair
TargetHistone-lysine N-methyltransferase EHMT2(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 15nMAssay Description:Inhibition G9a (unknown origin)More data for this Ligand-Target Pair
TargetHistone-lysine N-methyltransferase EHMT1(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 19nMAssay Description:Inhibition of GLP (unknown origin)More data for this Ligand-Target Pair
TargetHistone-lysine N-methyltransferase EHMT2(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 23nMAssay Description:Inhibition of recombinant human G9a using biotinylated-Histone H3 peptide (1 to 21 residues) after 30 mins in presence of SAM by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 25nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetHistone-lysine N-methyltransferase EHMT2(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 25nMAssay Description:Inhibition of recombinant human G9a using biotinylated-Histone H3 peptide (1 to 21 residues) after 30 mins in presence of SAM by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 30nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 40nMAssay Description:Demethylase reactions and AlphaScreen assays were performed as described (Sayegh et al., 2013), with a few exceptions. All demethylase buffers contai...More data for this Ligand-Target Pair
Affinity DataIC50: 59nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 60nMAssay Description:Irreversible inhibition of 0.4 uM KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 100 uM alpha-KG by succinate-glo ...More data for this Ligand-Target Pair
Affinity DataIC50: 61nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 67nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 74nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 100nMAssay Description:Demethylase reactions and AlphaScreen assays were performed as described (Sayegh et al., 2013), with a few exceptions. All demethylase buffers contai...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 100nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 10 uM alpha-KG by succinate-glo demethyl...More data for this Ligand-Target Pair
TargetHistone-lysine N-methyltransferase EHMT1(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 100nMAssay Description:Inhibition of GLP (unknown origin)More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 110nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using alpha-KG measured immediately by succina...More data for this Ligand-Target Pair
Affinity DataIC50: 110nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 110nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 100 uM alpha-KG by succinate-glo demethy...More data for this Ligand-Target Pair
Affinity DataIC50: 120nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 129nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 160nMAssay Description:Demethylase reactions and AlphaScreen assays were performed as described (Sayegh et al., 2013), with a few exceptions. All demethylase buffers contai...More data for this Ligand-Target Pair
TargetHistone-lysine N-methyltransferase EHMT2(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 160nMAssay Description:Inhibition G9a (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 175nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 177nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 180nMAssay Description:Irreversible inhibition of 0.8 uM KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 100 uM alpha-KG by succinate-glo ...More data for this Ligand-Target Pair
Affinity DataIC50: 180nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 180nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 1000 uM alpha-KG by succinate-glo demeth...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 220nMAssay Description:Irreversible inhibition of recombinant KDM5B ARID/PhD1 domain deletion mutant (1 to 755 residues) (unknown origin) expressed in Escherichia coli usin...More data for this Ligand-Target Pair
Affinity DataIC50: 240nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 290nMAssay Description:Inhibition of recombinant human C-terminal FLAG-tagged KDM5A (1 to 1090 residues) expressed in baculovirus infected sf9 cells using biotinylated H3K4...More data for this Ligand-Target Pair
Affinity DataIC50: 300nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
Affinity DataIC50: 303nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 320nMAssay Description:Irreversible inhibition of recombinant KDM5B ARID/PhD1 domain deletion mutant (1 to 755 residues) (unknown origin) expressed in Escherichia coli usin...More data for this Ligand-Target Pair
Affinity DataIC50: 330nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 360nMAssay Description:Irreversible inhibition of 1.6 uM KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 100 uM alpha-KG by succinate-glo ...More data for this Ligand-Target Pair
Affinity DataIC50: 370nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 400nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 430nMAssay Description:Inhibition of recombinant human C-terminal FLAG-tagged KDM5A (1 to 1090 residues) expressed in baculovirus infected sf9 cells using biotinylated H3K4...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 490nMAssay Description:Irreversible inhibition of recombinant KDM5B ARID/PhD1 domain deletion mutant (1 to 755 residues) (unknown origin) expressed in Escherichia coli usin...More data for this Ligand-Target Pair
Affinity DataIC50: 490nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 500nMAssay Description:Inhibition of ARID/PHD1 domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli using H3K4me3 as su...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 570nMAssay Description:Inhibition of recombinant human C-terminal FLAG-tagged KDM5A (1 to 1090 residues) expressed in baculovirus infected sf9 cells using biotinylated H3K4...More data for this Ligand-Target Pair
Affinity DataIC50: 580nMpH: 7.2 T: 2°CAssay Description:Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 590nMAssay Description:Inhibition of recombinant human C-terminal FLAG-tagged KDM5A (1 to 1090 residues) expressed in baculovirus infected sf9 cells using biotinylated H3K4...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 670nMAssay Description:Inhibition of recombinant human C-terminal FLAG-tagged KDM5A (1 to 1090 residues) expressed in baculovirus infected sf9 cells using biotinylated H3K4...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 690nMAssay Description:Inhibition of recombinant human C-terminal FLAG-tagged KDM5A (1 to 1090 residues) expressed in baculovirus infected sf9 cells using biotinylated H3K4...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataIC50: 700nMAssay Description:Irreversible inhibition of KDM5A ARID/PhD1 domain deletion mutant (1 to 739 residues) (unknown origin) using 10 uM alpha-KG by succinate-glo demethyl...More data for this Ligand-Target Pair


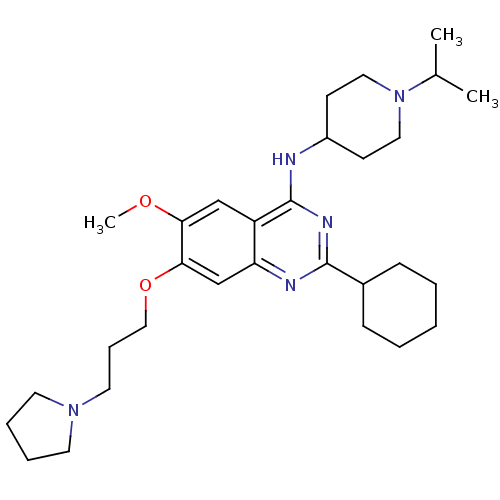
 3D Structure (crystal)
3D Structure (crystal)