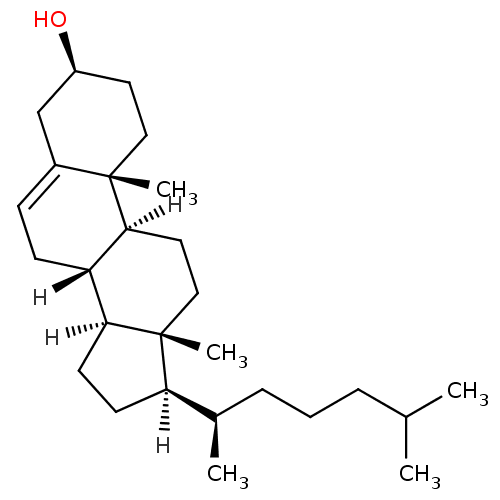
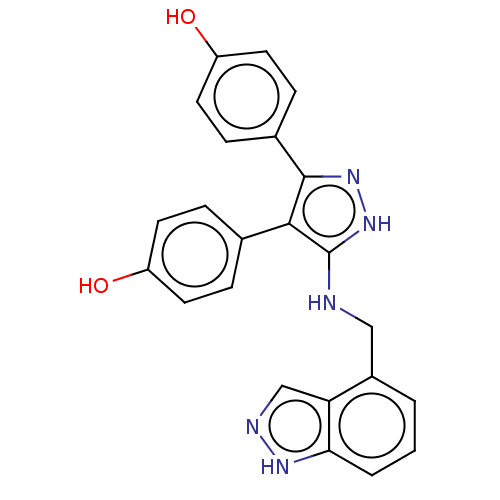
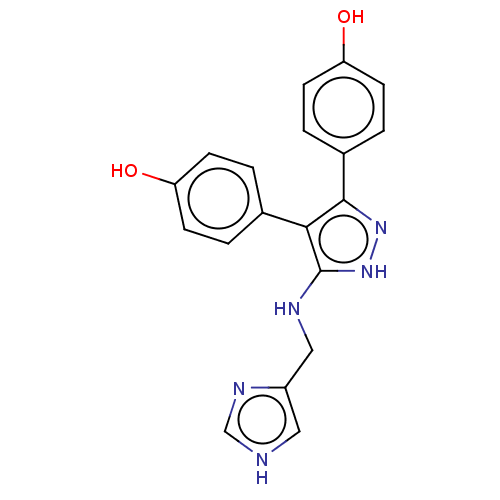
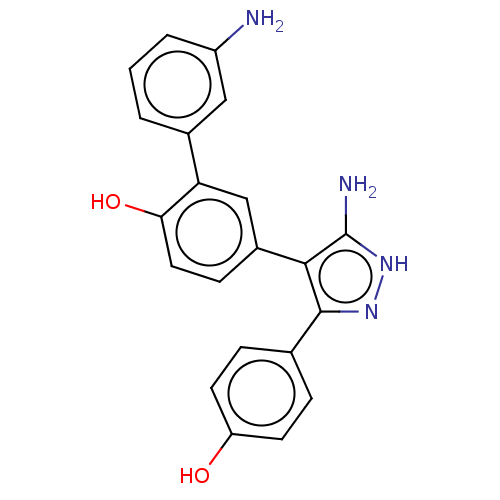
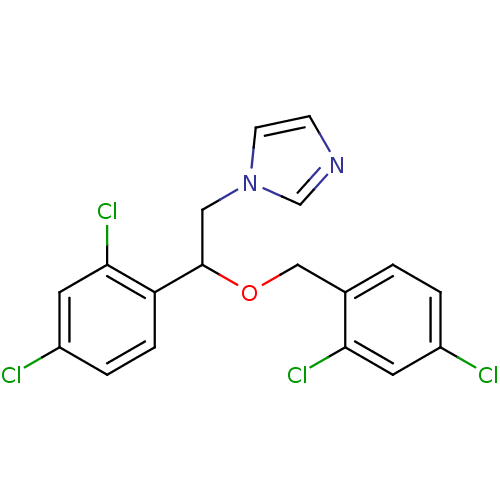
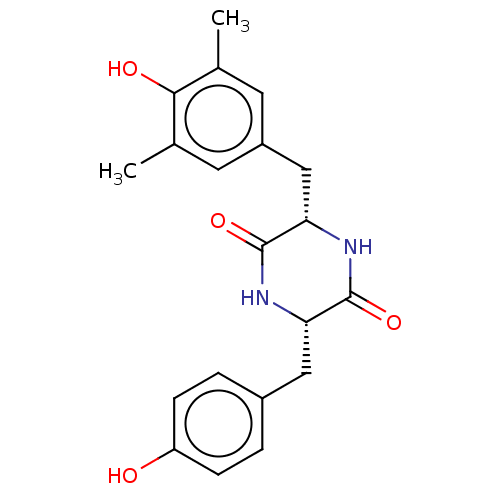
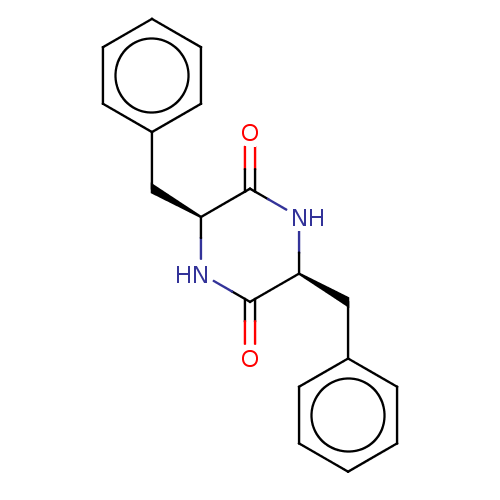
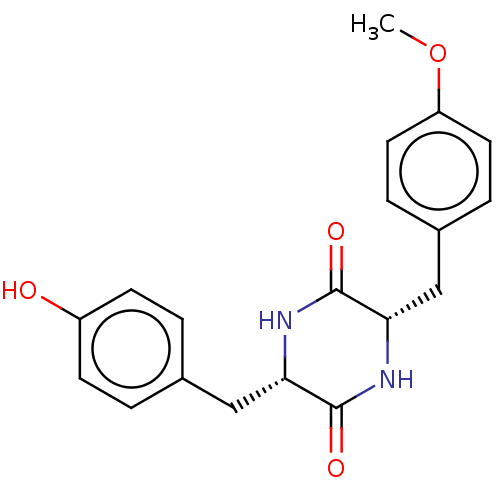
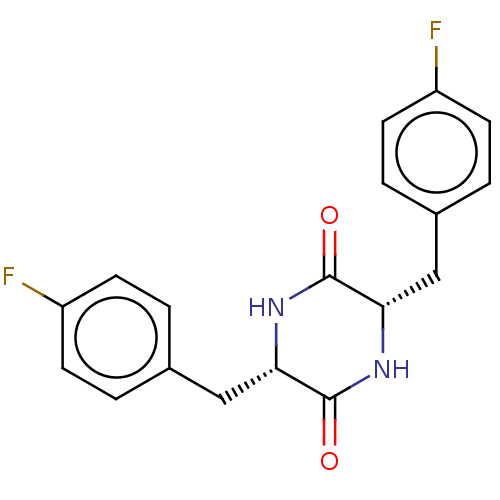
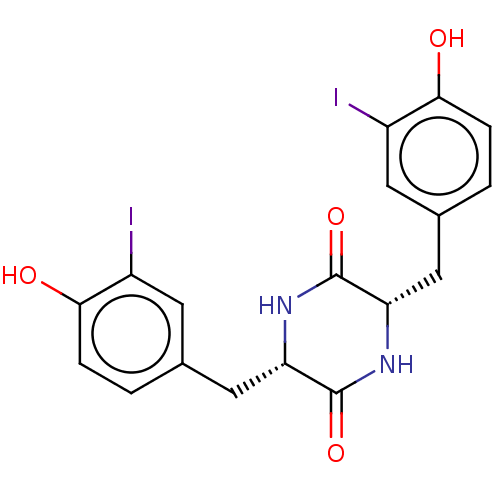
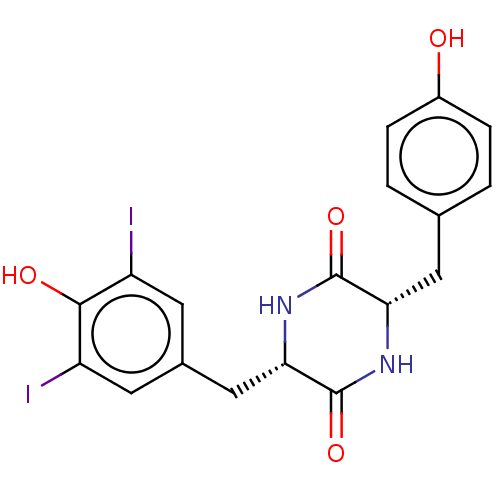
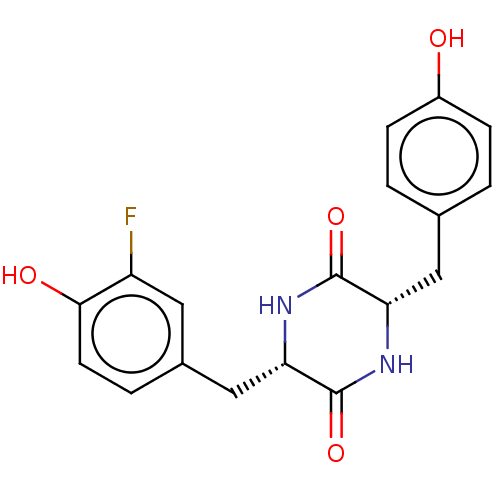
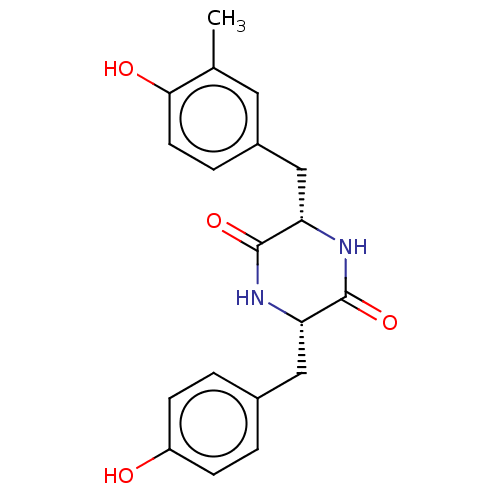
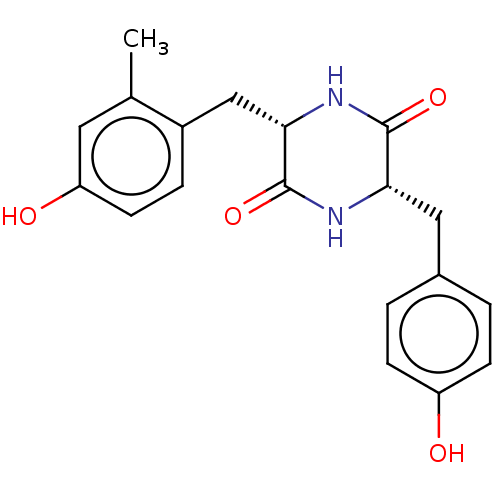
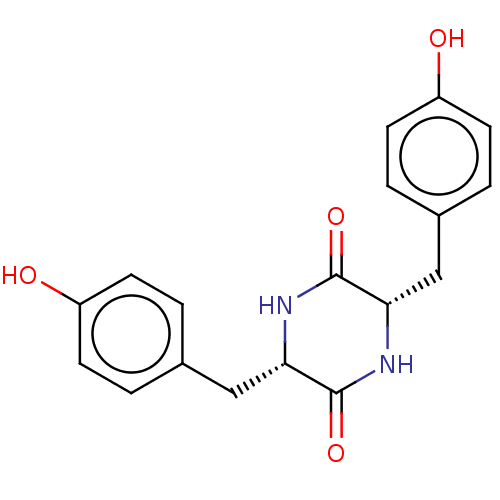
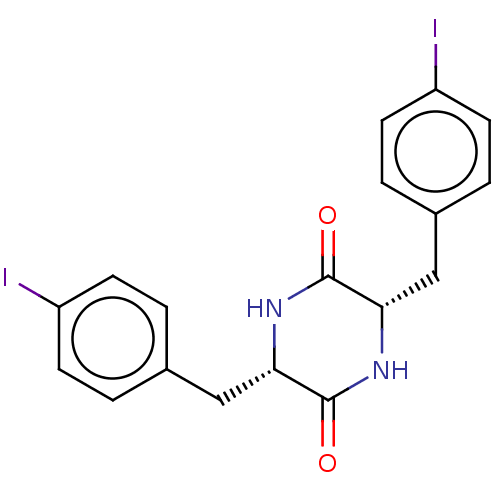
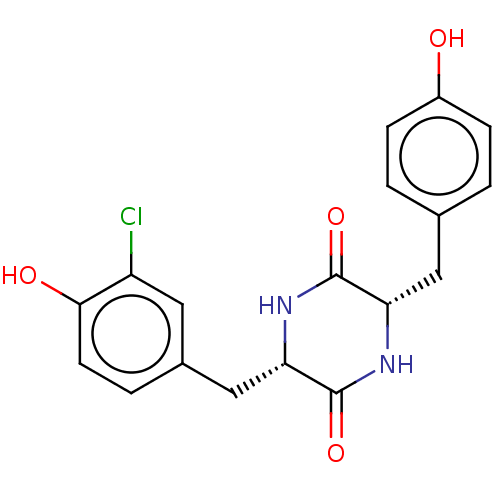
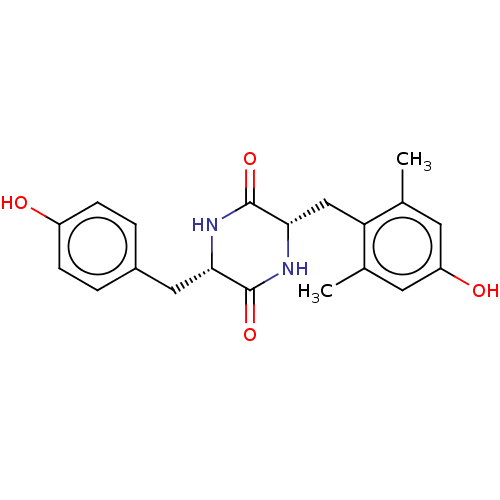
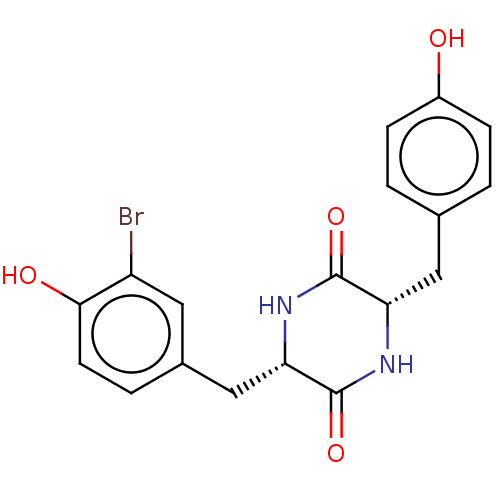
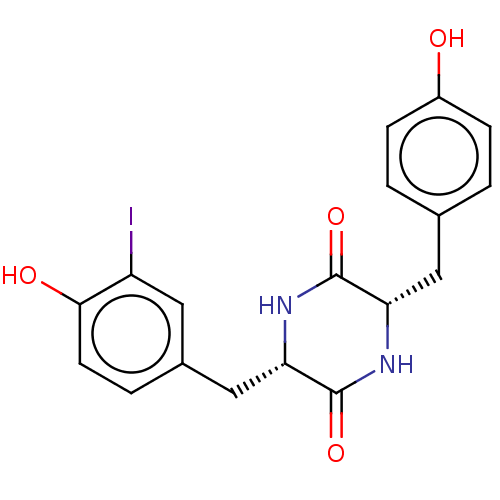
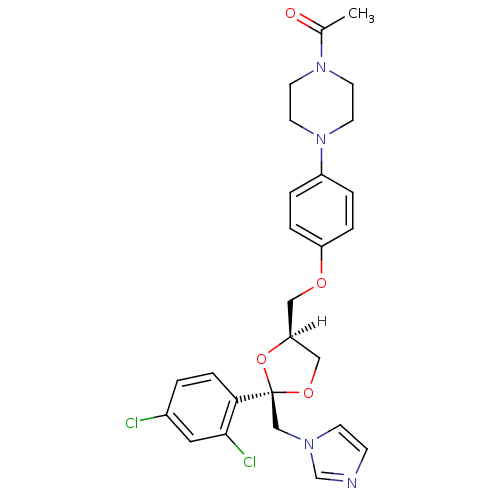
Affinity DataIC50: 1.13E+4nMAssay Description:Inhibition of CYP3A4 in human liver microsomes using midazolam as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2C19 in human liver microsomes using mephenytoin as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2C19 in human liver microsomes using mephenytoin as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2C19 in human liver microsomes using mephenytoin as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2D6 in human liver microsomes using dextromethorphan as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2D6 in human liver microsomes using dextromethorphan as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2C9 in human liver microsomes using tolbutamide as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2C9 in human liver microsomes using tolbutamide as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2C9 in human liver microsomes using tolbutamide as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP3A4 in human liver microsomes using midazolam as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2D6 in human liver microsomes using dextromethorphan as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP3A4 in human liver microsomes using midazolam as substrate by LC-MS/MS analysisMore data for this Ligand-Target Pair
Affinity DataKd: 4.31E+4nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 8.60E+5nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 1.74E+5nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 3.02E+4nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 2.80E+5nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 2.16E+5nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 1.20E+4nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 1.20E+3nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 360nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 110nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 340nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 8.20E+4nMpH: 7.0Assay Description:Optical titrations to determine the binding affinity (KD values) of ligands were carried out on a Cary 60 UV−visible spectrophotometer (Varian,...More data for this Ligand-Target Pair
Affinity DataKd: 2.00E+5nMpH: 7.0Assay Description:Optical titrations to determine the binding affinity (KD values) of ligands were carried out on a Cary 60 UV−visible spectrophotometer (Varian,...More data for this Ligand-Target Pair
Affinity DataKd: 1.40E+5nMpH: 7.5 T: 2°CAssay Description:ITC binding isotherms were recorded on a MicroCal ITC200 instrument (MalvernInstruments). Titrations were conducted at 25 °C by injecting aliquots (2...More data for this Ligand-Target Pair
Affinity DataKd: 2.98E+6nMpH: 7.0Assay Description:Optical titrations to determine the binding affinity (KD values) of ligands were carried out on a Cary 60 UV−visible spectrophotometer (Varian,...More data for this Ligand-Target Pair
Affinity DataKd: 2.90E+5nMpH: 7.0Assay Description:Optical titrations to determine the binding affinity (KD values) of ligands were carried out on a Cary 60 UV−visible spectrophotometer (Varian,...More data for this Ligand-Target Pair
Affinity DataKd: 2.30E+5nMpH: 7.0Assay Description:Optical titrations to determine the binding affinity (KD values) of ligands were carried out on a Cary 60 UV−visible spectrophotometer (Varian,...More data for this Ligand-Target Pair
Affinity DataKd: 73nMAssay Description:Binding affinity to Mycobacterium tuberculosis H37Rv wild type CYP121 by titration assayMore data for this Ligand-Target Pair
Affinity DataKd: 73nMAssay Description:Binding affinity to Mycobacterium tuberculosis H37Rv wild type CYP121 by titration assayMore data for this Ligand-Target Pair
Affinity DataKd: 73nMAssay Description:Binding affinity to Mycobacterium tuberculosis H37Rv wild type CYP121 by titration assayMore data for this Ligand-Target Pair
Affinity DataKd: 2.94E+4nMAssay Description:Binding affinity to Mycobacterium tuberculosis CYP121 expressed in Escherichia coli HMS174 (DE3) by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.51E+4nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 2.40E+3nMAssay Description:Binding affinity to Mycobacterium tuberculosis CYP121 expressed in Escherichia coli HMS174 (DE3) by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.04E+4nMAssay Description:Binding affinity to Mycobacterium tuberculosis CYP121 expressed in Escherichia coli HMS174 (DE3) by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 6.30E+3nMAssay Description:Binding affinity to Mycobacterium tuberculosis CYP121 expressed in Escherichia coli HMS174 (DE3) by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 450nMAssay Description:Binding affinity to Mycobacterium tuberculosis CYP121 expressed in Escherichia coli HMS174 (DE3) by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 9.10E+3nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.28E+4nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.10E+3nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 3.98E+4nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 3.00E+4nMAssay Description:Binding affinity to Mycobacterium tuberculosis CYP121 expressed in Escherichia coli HMS174 (DE3) by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 720nMAssay Description:Binding affinity to Mycobacterium tuberculosis CYP121 expressed in Escherichia coli HMS174 (DE3) by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.50E+3nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: >1.00E+5nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 460nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 280nMAssay Description:Binding affinity to Mycobacterium tuberculosis His6-tagged CYP121 by UV-visible scanning spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataKd: 2.10E+4nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair
Affinity DataKd: 2.71E+4nMAssay Description:Substrate and ligand binding assay using UV- visible absorbance analysis of CYP142 was done on a Cary UV-50 UV-visible scanning spectrophotometer (Va...More data for this Ligand-Target Pair

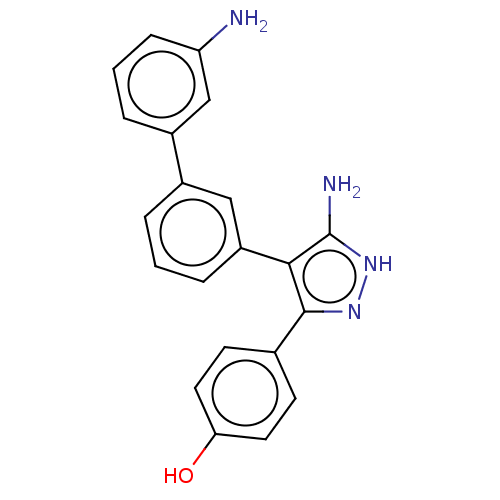
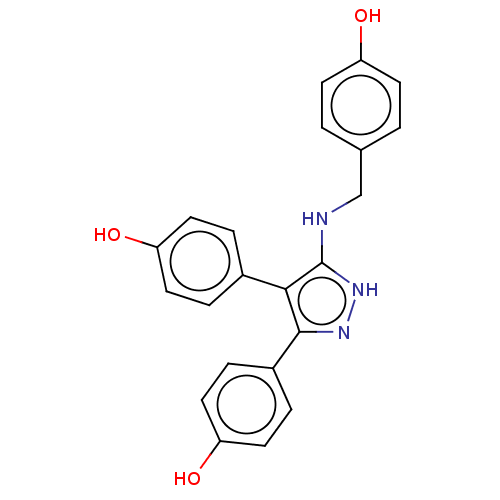
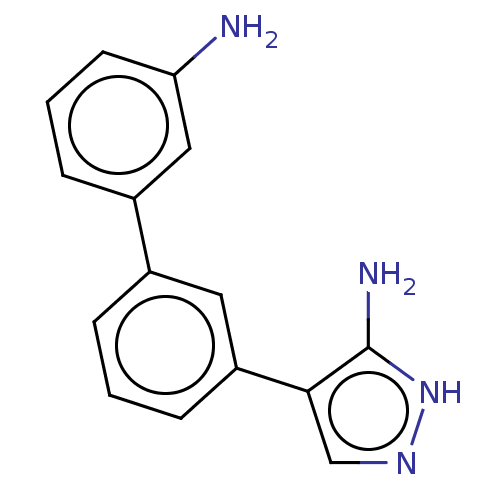
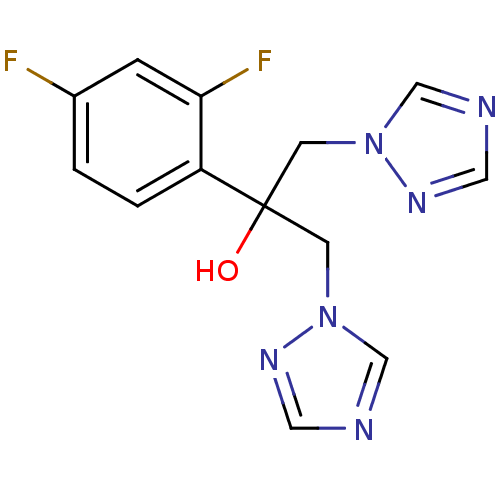
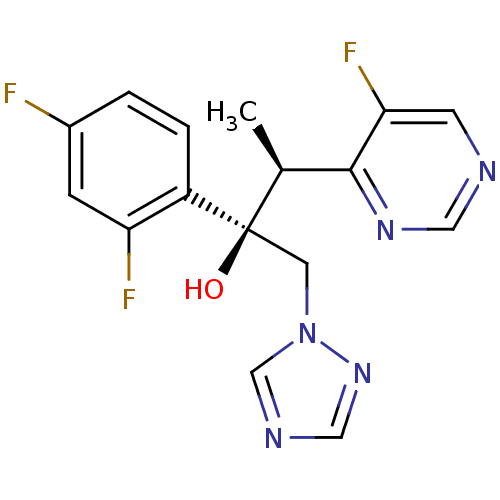
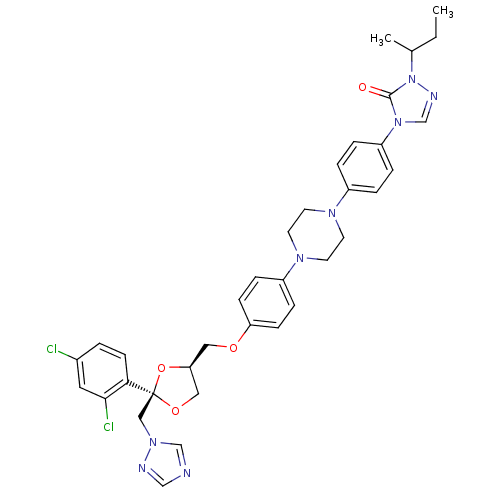
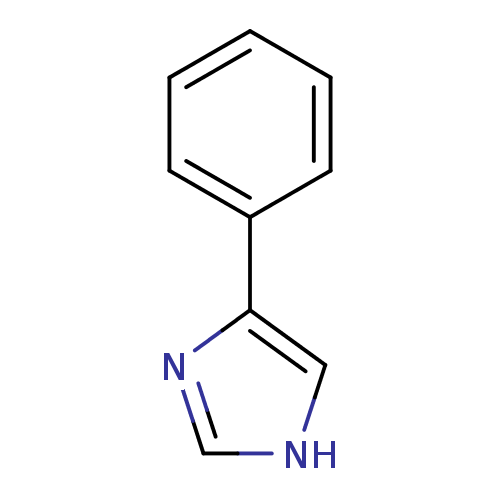

 3D Structure (crystal)
3D Structure (crystal)