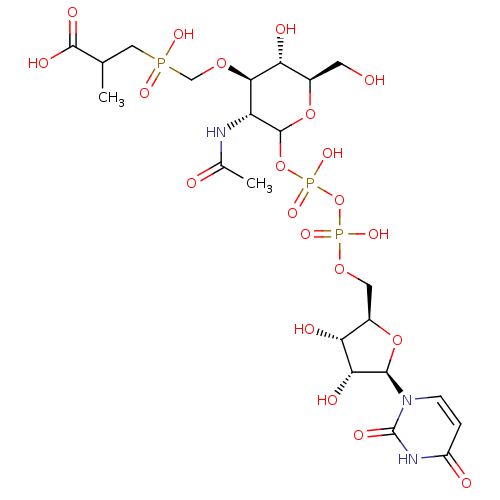
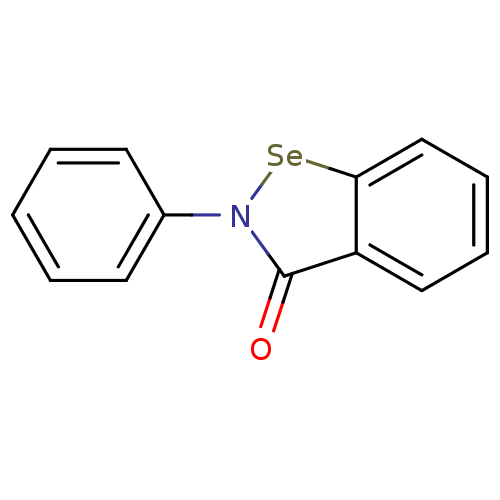
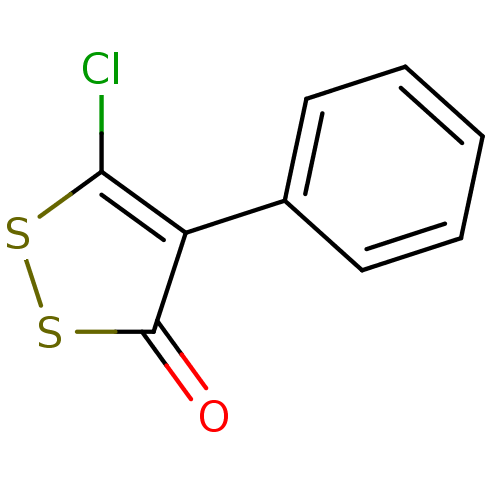
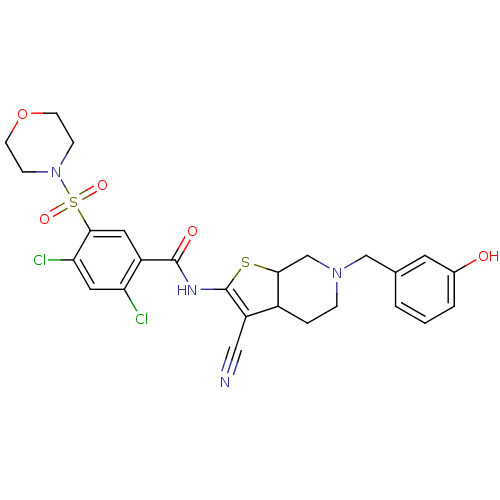
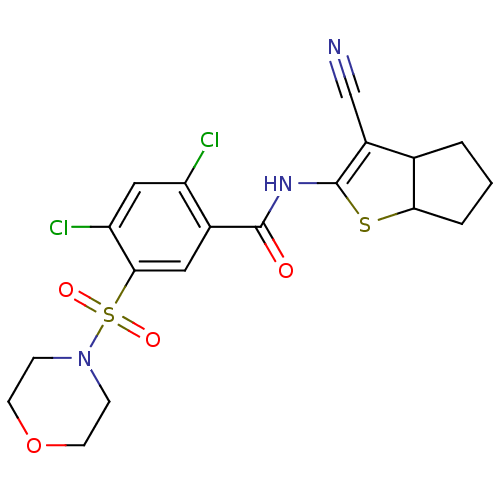
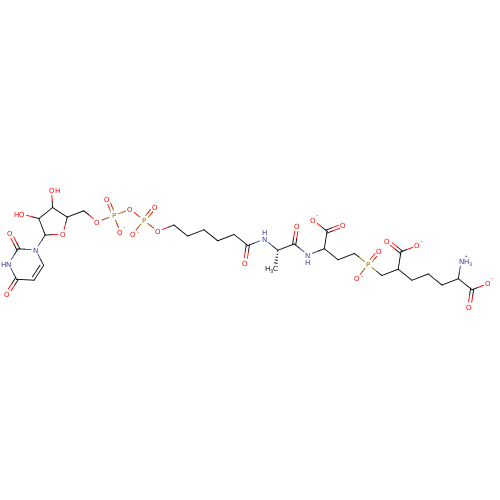
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase 2(Staphylococcus aureus (Firmicutes))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Enterobacter cloacae (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
TargetUDP-N-acetylmuramoylalanine--D-glutamate ligase(Escherichia coli (strain K12))
Freie Universit£t Berlin
Curated by ChEMBL
Freie Universit£t Berlin
Curated by ChEMBL
Affinity DataKi: 2.10E+4nMAssay Description:Competitive inhibition of Escherichia coli MurD using D-Glu substrate by steady-state enzyme kinetics assayMore data for this Ligand-Target Pair
TargetUDP-N-acetylmuramoylalanine--D-glutamate ligase(Escherichia coli (strain K12))
Freie Universit£t Berlin
Curated by ChEMBL
Freie Universit£t Berlin
Curated by ChEMBL
Affinity DataKi: 2.90E+4nMAssay Description:Partial non-competitive inhibition of Escherichia coli MurD using UMA substrate by steady-state enzyme kinetics assayMore data for this Ligand-Target Pair
TargetUDP-N-acetylmuramoylalanine--D-glutamate ligase(Escherichia coli (strain K12))
Freie Universit£t Berlin
Curated by ChEMBL
Freie Universit£t Berlin
Curated by ChEMBL
Affinity DataKi: 3.90E+4nMAssay Description:Partial un-competitive inhibition of Escherichia coli MurD using ATP substrate by steady-state enzyme kinetics assayMore data for this Ligand-Target Pair
TargetUDP-N-acetylmuramoylalanine--D-glutamate ligase(Escherichia coli (strain K12))
Freie Universit£t Berlin
Curated by ChEMBL
Freie Universit£t Berlin
Curated by ChEMBL
Affinity DataIC50: 4nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 6nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 7nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 9nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 9nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 9nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 14nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 14nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
TargetUDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase(Escherichia coli)
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 26nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 35nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
TargetUDP-N-acetylmuramate--L-alanine ligase(Escherichia coli)
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 50nMAssay Description:Inhibition of Staphylococcus aureus topoisomerase 4 assessed as reduction in decatenation using supercoiled pNO1 plasmid DNA as substrate incubated f...More data for this Ligand-Target Pair
Affinity DataIC50: 67nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 87nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 90nMAssay Description:Inhibition of Staphylococcus aureus topoisomerase 4 assessed as reduction in decatenation using supercoiled pNO1 plasmid DNA as substrate incubated f...More data for this Ligand-Target Pair
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Haemophilus influenzae (G-proteobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 123nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 126nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 172nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 197nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 215nMAssay Description:Inhibition of Staphylococcus aureus topoisomerase 4 assessed as reduction in decatenation using supercoiled pNO1 plasmid DNA as substrate incubated f...More data for this Ligand-Target Pair
Affinity DataIC50: 236nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 241nMAssay Description:Inhibition of Staphylococcus aureus topoisomerase 4 assessed as reduction in decatenation using supercoiled pNO1 plasmid DNA as substrate incubated f...More data for this Ligand-Target Pair
Affinity DataIC50: 244nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 280nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 283nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
Affinity DataIC50: 326nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 356nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 374nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30...More data for this Ligand-Target Pair
TargetUDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase(Streptococcus pneumoniae (Firmicutes))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 570nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 725nMAssay Description:Inhibition of Staphylococcus aureus topoisomerase 4 assessed as reduction in decatenation using supercoiled pNO1 plasmid DNA as substrate incubated f...More data for this Ligand-Target Pair
TargetUDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase(Escherichia coli)
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 1.02E+3nMAssay Description:Inhibition of Staphylococcus aureus topoisomerase 4 assessed as reduction in decatenation using supercoiled pNO1 plasmid DNA as substrate incubated f...More data for this Ligand-Target Pair
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase 2(Staphylococcus aureus (Firmicutes))
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
TargetUDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L-lysine ligase(Staphylococcus aureus)
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia
Affinity DataIC50: 1.24E+3nMAssay Description:Inhibition of Staphylococcus aureus topoisomerase 4 assessed as reduction in decatenation using supercoiled pNO1 plasmid DNA as substrate incubated f...More data for this Ligand-Target Pair
Affinity DataIC50: 1.71E+3nMAssay Description:Inhibition of Escherichia coli DNA gyrase assessed as reduction in DNA supercoiling using relaxed pNO1 plasmid DNA as substrate incubated for 30 mins...More data for this Ligand-Target Pair
Affinity DataIC50: 1.88E+3nMAssay Description:Inhibition of Staphylococcus aureus topoisomerase 4 assessed as reduction in decatenation using supercoiled pNO1 plasmid DNA as substrate incubated f...More data for this Ligand-Target Pair

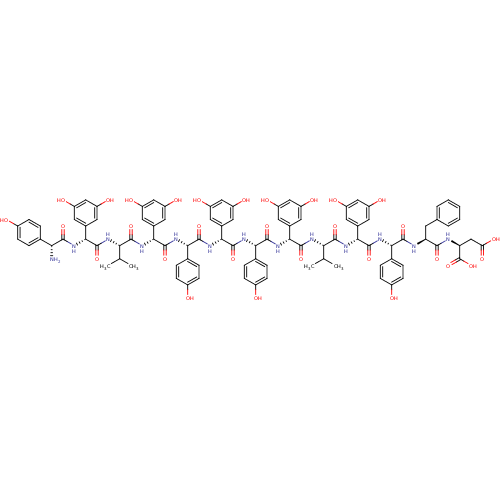
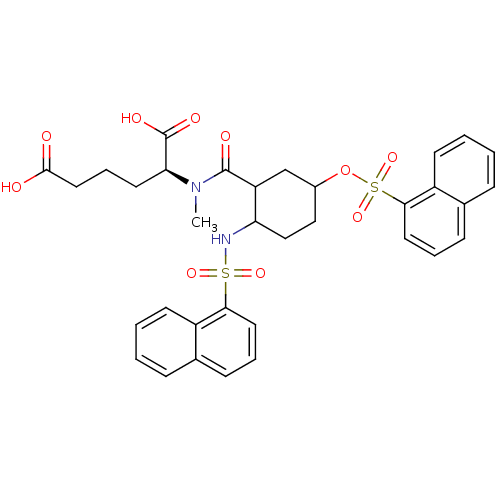
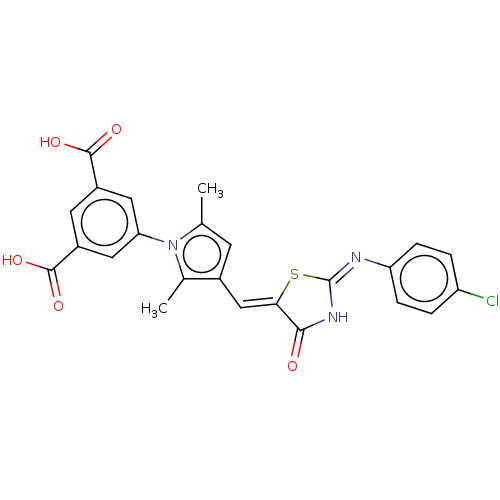
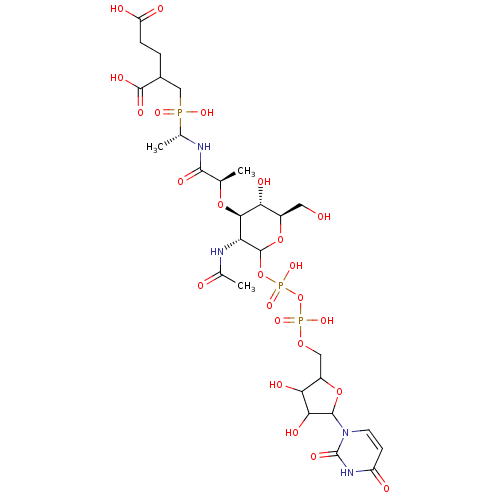
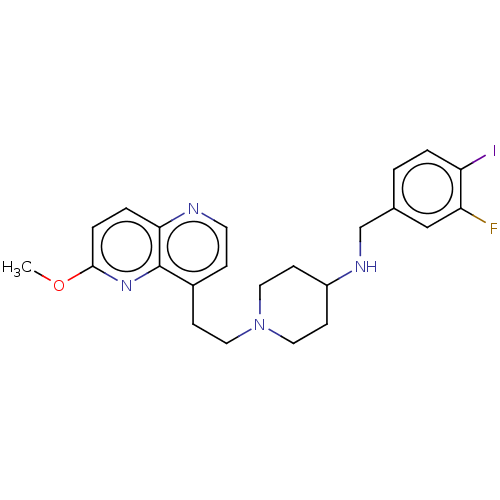

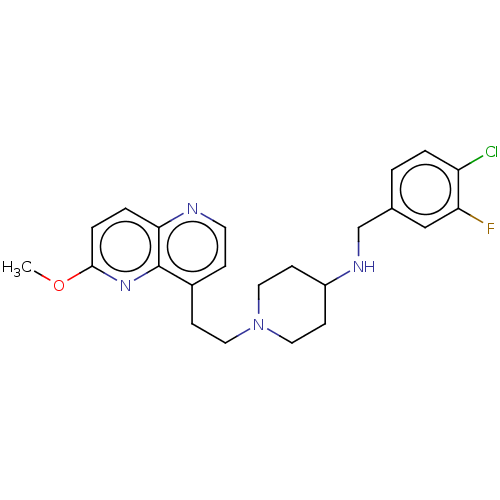







 3D Structure (crystal)
3D Structure (crystal)