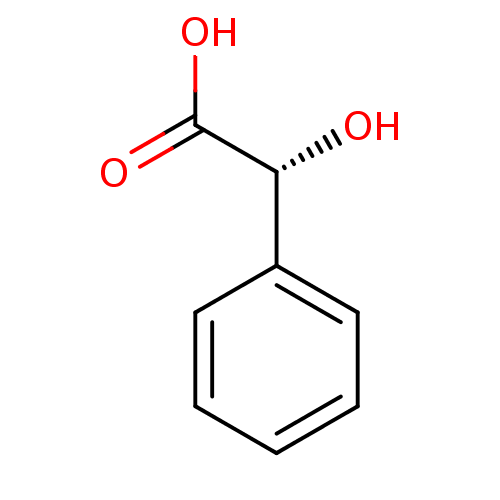
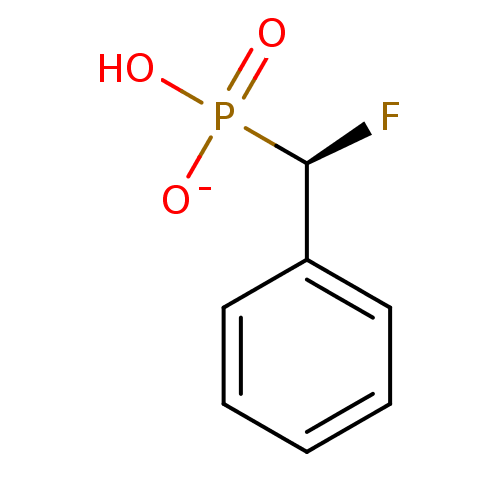
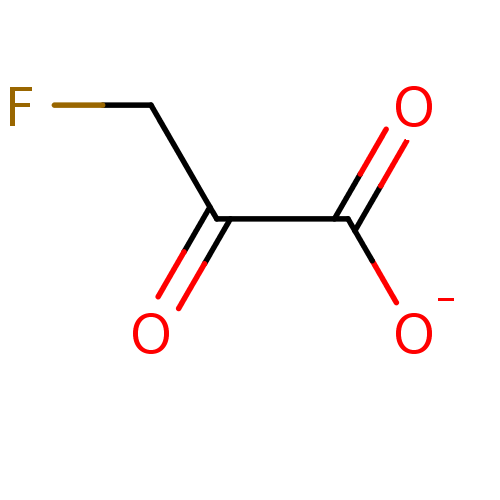
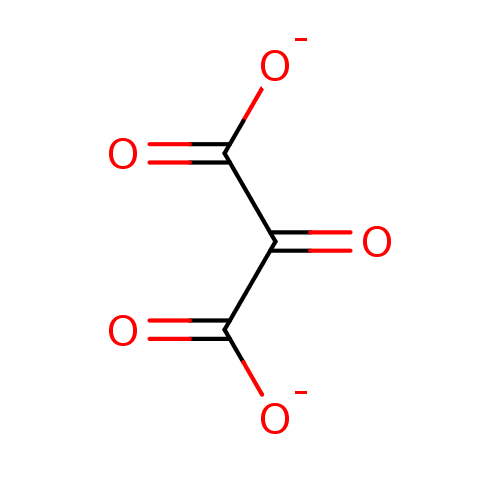
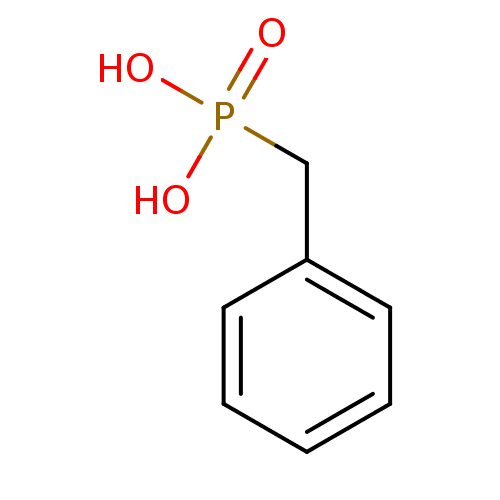
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 1.10E+3nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 4.70E+3nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 3.40E+4nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 3.00E+5nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 5.30E+5nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 5.70E+5nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 5.90E+5nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 8.10E+5nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 1.11E+6nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 1.30E+6nMpH: 7.5Assay Description:MR activity was assayed using a CD-based assay by following the change in ellipticity of mandelate at 262 nm using a thermostated quartz cuvette with...More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 1.80E+6nMpH: 7.5Assay Description:MR activity was assayed using a CD-based assay by following the change in ellipticity of mandelate at 262 nm using a thermostated quartz cuvette with...More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 3.60E+6nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair
TargetMandelate racemase(Pseudomonas putida (g-Proteobacteria))
Dalhousie University
Curated by ChEMBL
Dalhousie University
Curated by ChEMBL
Affinity DataKi: 1.20E+7nMpH: 7.5Assay Description:Inhibition constant against Mandelate racemase from Pseudomonas putida at pH 7.5More data for this Ligand-Target Pair

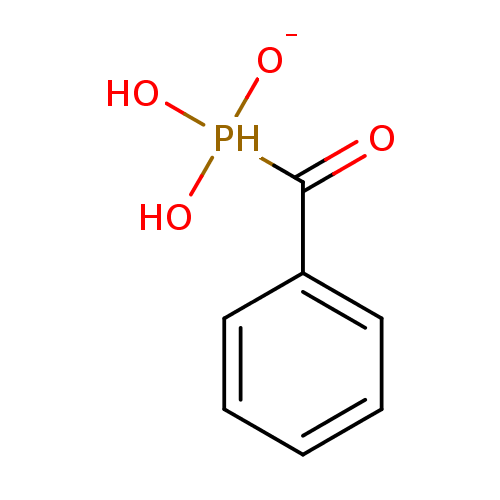


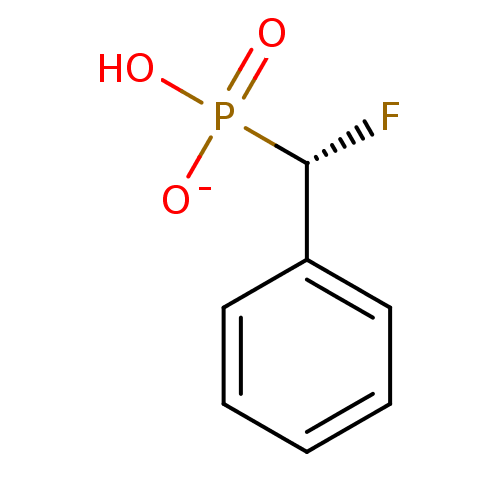
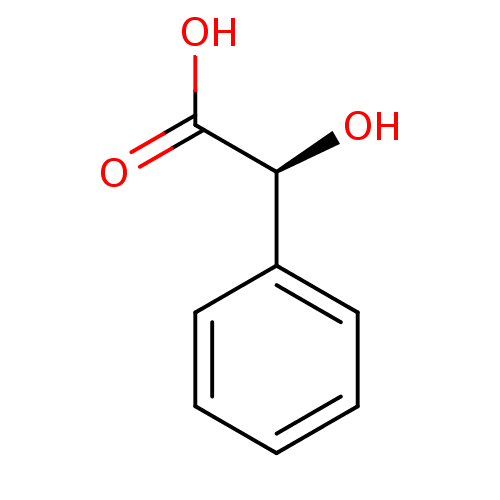
 3D Structure (crystal)
3D Structure (crystal)