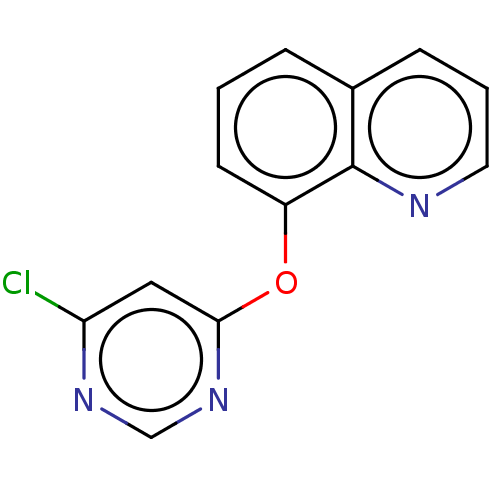
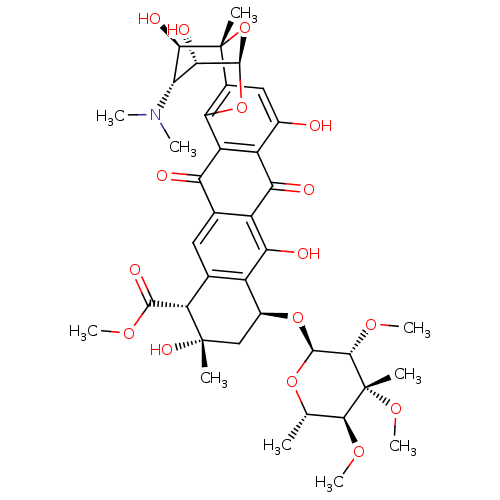
TargetCalcium/calmodulin-dependent protein kinase type IV [15-340](Homo sapiens (Human))
B.R. Ambedkar Bihar University
B.R. Ambedkar Bihar University
Affinity DataKi: 0.0109nMAssay Description:The docking and scoring of ligands with CAMKIV protein was accomplished using ParDOCK module of Sanjeevini drug design suite, which is based on physi...More data for this Ligand-Target Pair
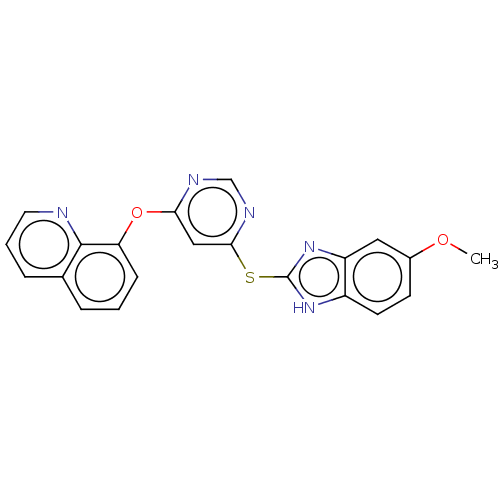
TargetCalcium/calmodulin-dependent protein kinase type IV [15-340](Homo sapiens (Human))
B.R. Ambedkar Bihar University
B.R. Ambedkar Bihar University
Affinity DataKi: 1.35E+3nMAssay Description:The docking and scoring of ligands with CAMKIV protein was accomplished using ParDOCK module of Sanjeevini drug design suite, which is based on physi...More data for this Ligand-Target Pair
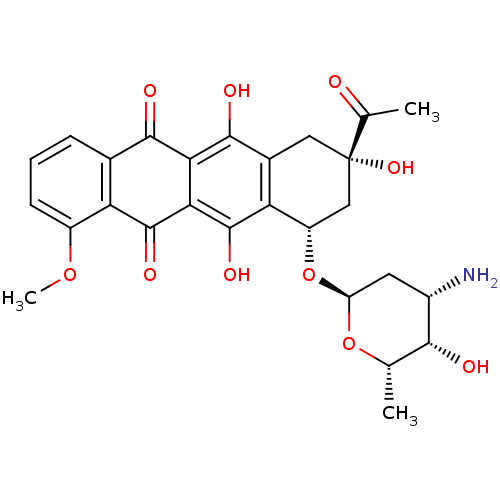
TargetCalcium/calmodulin-dependent protein kinase type IV [15-340](Homo sapiens (Human))
B.R. Ambedkar Bihar University
B.R. Ambedkar Bihar University
Affinity DataKi: 1.45E+3nMAssay Description:The docking and scoring of ligands with CAMKIV protein was accomplished using ParDOCK module of Sanjeevini drug design suite, which is based on physi...More data for this Ligand-Target Pair
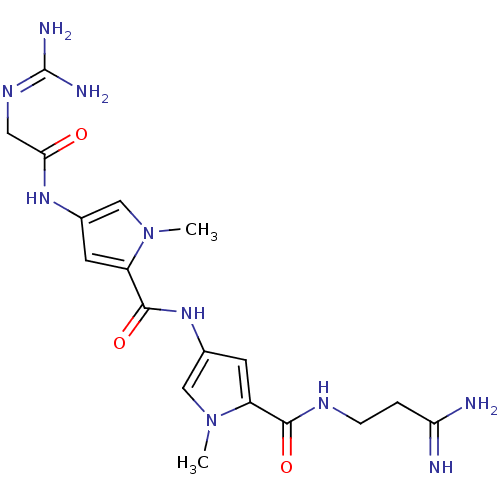
TargetCalcium/calmodulin-dependent protein kinase type IV [15-340](Homo sapiens (Human))
B.R. Ambedkar Bihar University
B.R. Ambedkar Bihar University
Affinity DataKi: 3.57E+3nMAssay Description:The docking and scoring of ligands with CAMKIV protein was accomplished using ParDOCK module of Sanjeevini drug design suite, which is based on physi...More data for this Ligand-Target Pair
TargetCalcium/calmodulin-dependent protein kinase type IV [15-340](Homo sapiens (Human))
B.R. Ambedkar Bihar University
B.R. Ambedkar Bihar University
Affinity DataKi: 1.01E+4nMAssay Description:The docking and scoring of ligands with CAMKIV protein was accomplished using ParDOCK module of Sanjeevini drug design suite, which is based on physi...More data for this Ligand-Target Pair
TargetCalcium/calmodulin-dependent protein kinase type IV [15-340](Homo sapiens (Human))
B.R. Ambedkar Bihar University
B.R. Ambedkar Bihar University
Affinity DataKi: 2.44E+4nMAssay Description:The docking and scoring of ligands with CAMKIV protein was accomplished using ParDOCK module of Sanjeevini drug design suite, which is based on physi...More data for this Ligand-Target Pair
TargetCalcium/calmodulin-dependent protein kinase type IV [15-340](Homo sapiens (Human))
B.R. Ambedkar Bihar University
B.R. Ambedkar Bihar University
Affinity DataKi: 6.67E+5nMAssay Description:The docking and scoring of ligands with CAMKIV protein was accomplished using ParDOCK module of Sanjeevini drug design suite, which is based on physi...More data for this Ligand-Target Pair
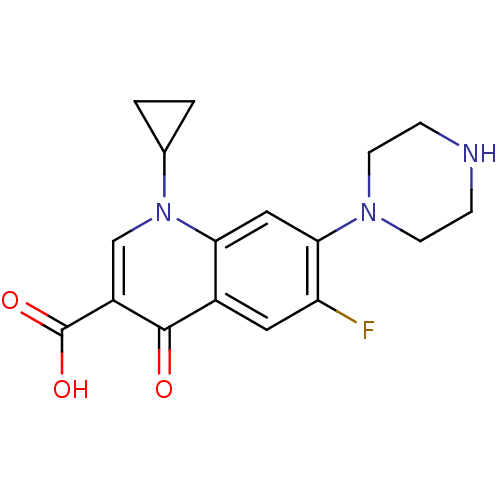
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 3.00E+3nMpH: 8.0 T: 2°CAssay Description:Helicase assay was demonstrated using the purified fraction of PfUDN. The specially designed partial duplex substrate consisted of a 32P-labelled 47-...More data for this Ligand-Target Pair
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 3.10E+3nMpH: 8.0 T: 2°CAssay Description:The hydrolysis of ATP catalyzed by PfUDN was assayed by measuring the formation of Pi from [γ-32P] ATP. The reaction mixture of 10 μl conta...More data for this Ligand-Target Pair
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 3.10E+3nMpH: 8.0 T: 2°CAssay Description:The hydrolysis of ATP catalyzed by PfUDN was assayed by measuring the formation of Pi from [γ-32P] ATP. The reaction mixture of 10 μl conta...More data for this Ligand-Target Pair
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 3.30E+3nMpH: 8.0 T: 2°CAssay Description:The hydrolysis of ATP catalyzed by PfUDN was assayed by measuring the formation of Pi from [γ-32P] ATP. The reaction mixture of 10 μl conta...More data for this Ligand-Target Pair
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 3.50E+3nMpH: 8.0 T: 2°CAssay Description:Helicase assay was demonstrated using the purified fraction of PfUDN. The specially designed partial duplex substrate consisted of a 32P-labelled 47-...More data for this Ligand-Target Pair
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 4.40E+3nMpH: 8.0 T: 2°CAssay Description:The hydrolysis of ATP catalyzed by PfUDN was assayed by measuring the formation of Pi from [γ-32P] ATP. The reaction mixture of 10 μl conta...More data for this Ligand-Target Pair
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 4.70E+3nMpH: 8.0 T: 2°CAssay Description:Helicase assay was demonstrated using the purified fraction of PfUDN. The specially designed partial duplex substrate consisted of a 32P-labelled 47-...More data for this Ligand-Target Pair
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 4.80E+3nMpH: 8.0 T: 2°CAssay Description:Helicase assay was demonstrated using the purified fraction of PfUDN. The specially designed partial duplex substrate consisted of a 32P-labelled 47-...More data for this Ligand-Target Pair
TargetDNA helicase [35-727](Plasmodium falciparum)
International Centre for Genetic Engineering and Biotechnology
International Centre for Genetic Engineering and Biotechnology
Affinity DataIC50: 9.90E+3nMpH: 8.0 T: 2°CAssay Description:The hydrolysis of ATP catalyzed by PfUDN was assayed by measuring the formation of Pi from [γ-32P] ATP. The reaction mixture of 10 μl conta...More data for this Ligand-Target Pair