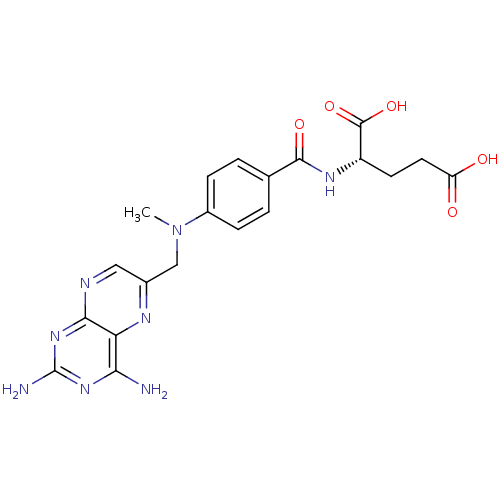
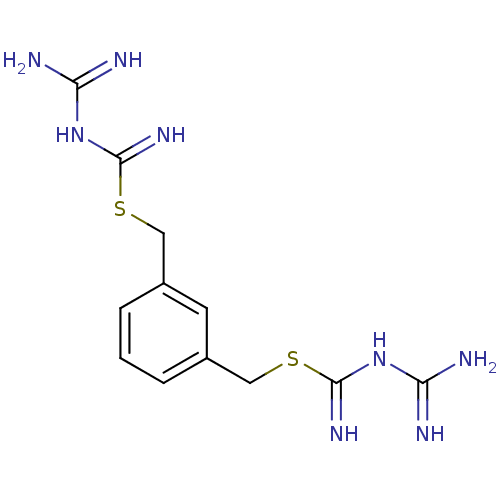
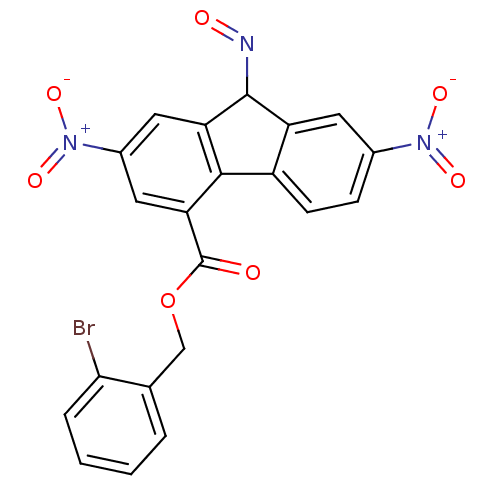
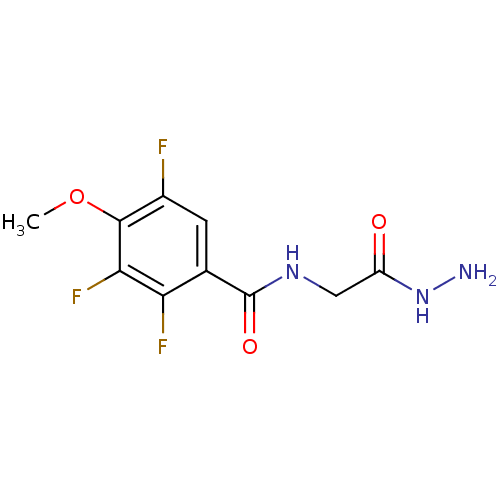
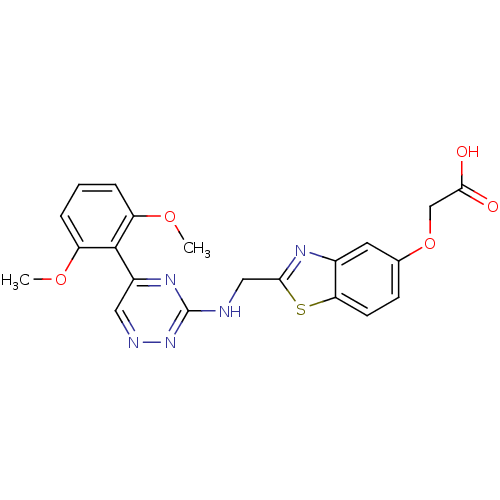
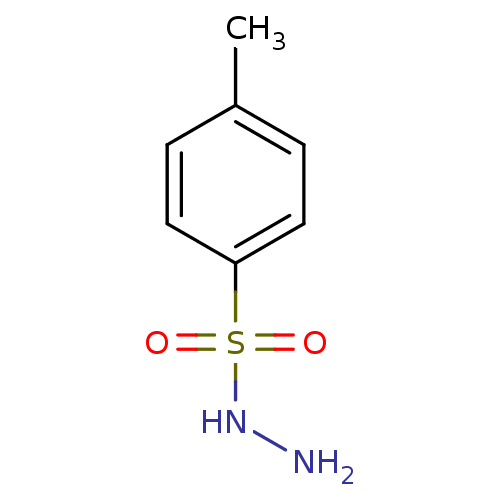
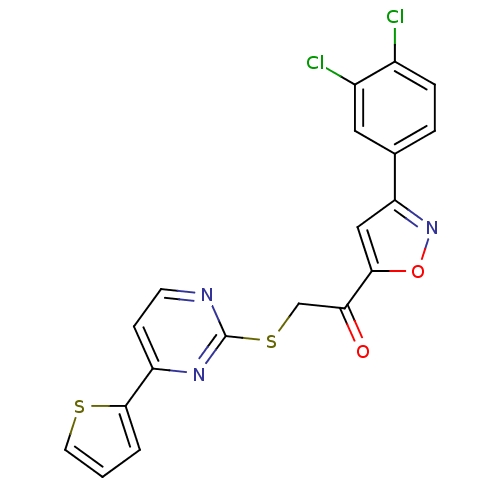
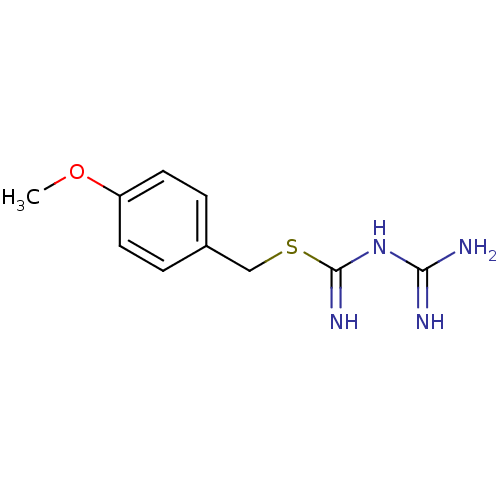
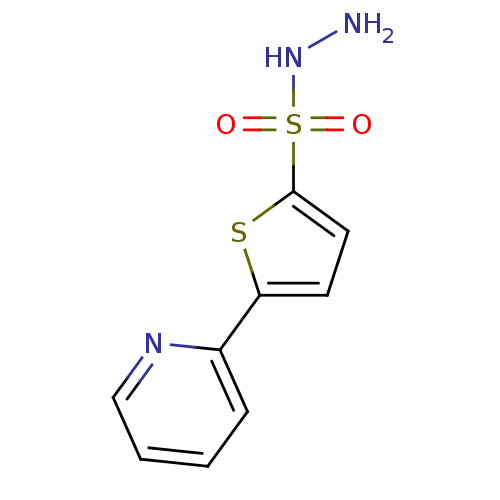
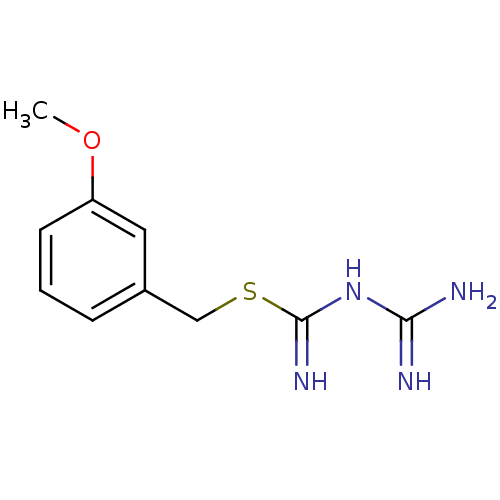
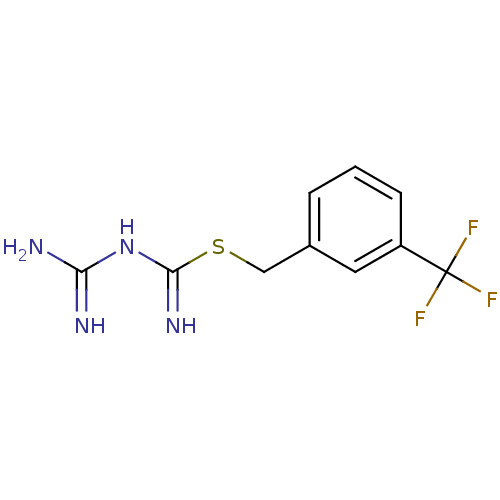
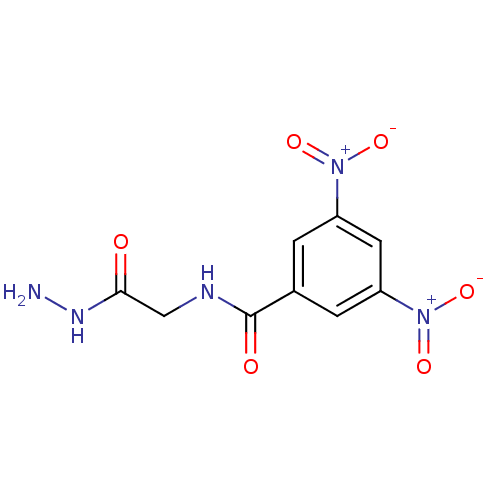
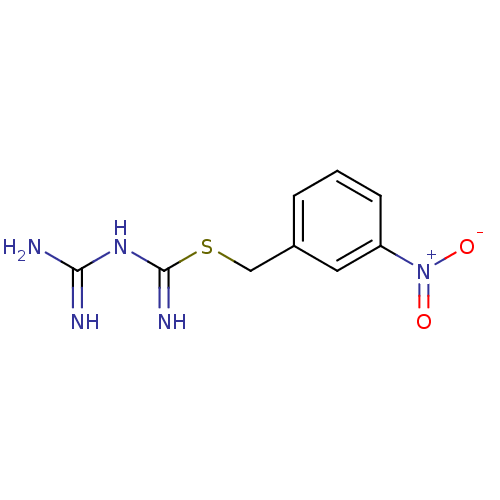
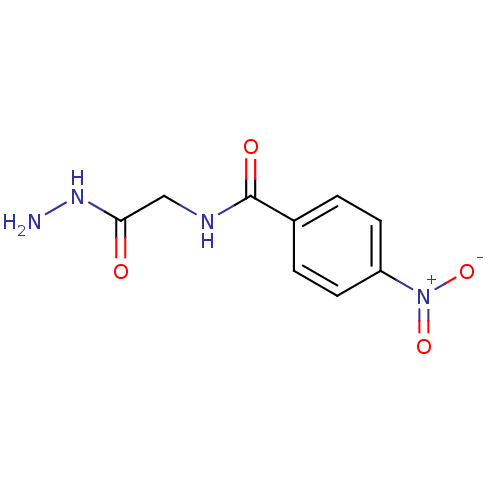
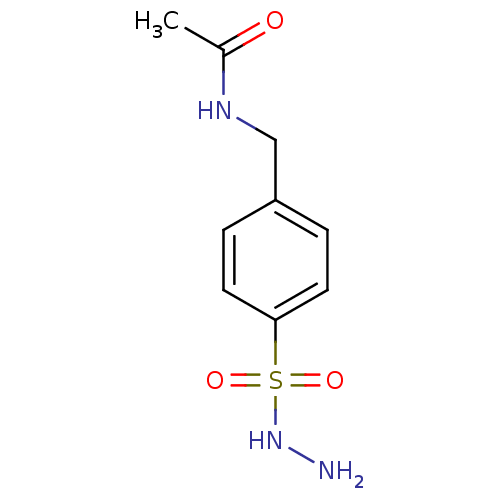
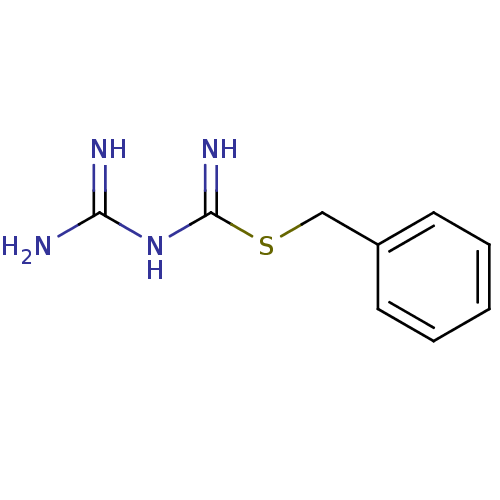
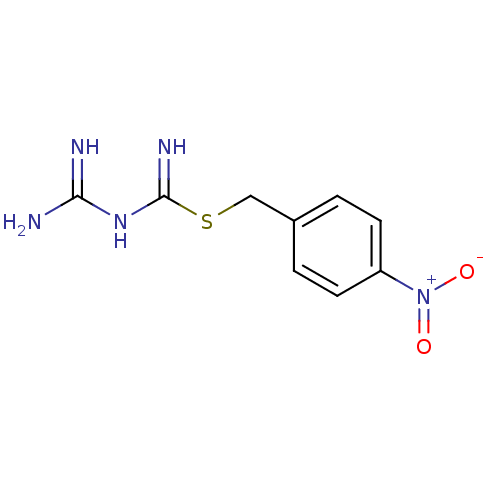
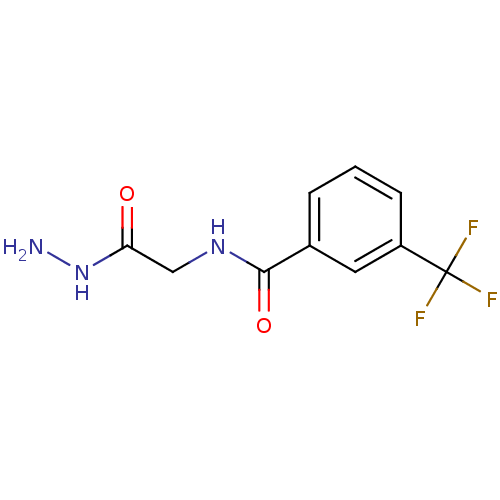
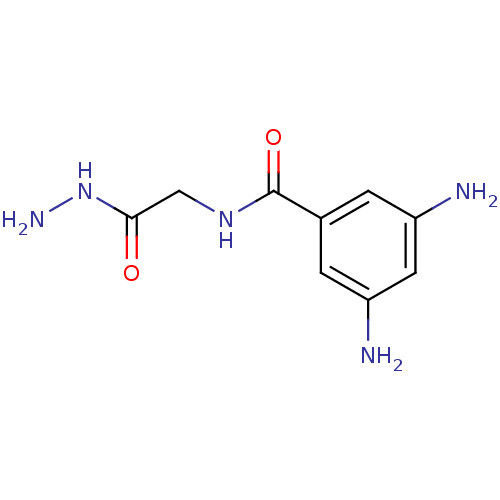
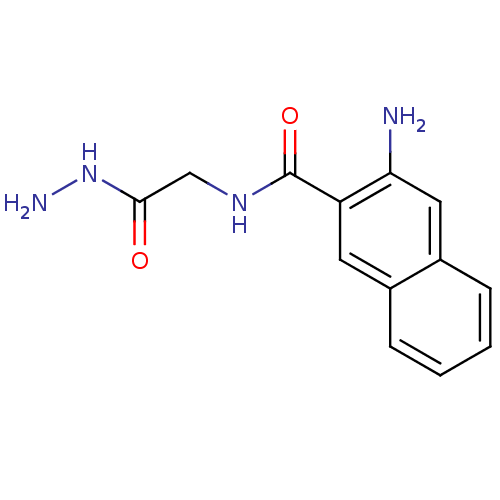
Affinity DataKi: 11.5nM ΔG°: -45.3kJ/molepH: 7.5 T: 2°CAssay Description:The oxidation of NADPH was monitored at 340 nm using a Molecular Devices SpectraMax Plus 96-well microtiter plate reading spectrophotometer. Plots we...More data for this Ligand-Target Pair
Affinity DataIC50: 3nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 75nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 75nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 79nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
TargetD-beta-D-heptose 7-phosphate kinase(Burkholderia cepacia (strain J2315 / LMG 16656) (B...)
McMaster University
Curated by ChEMBL
McMaster University
Curated by ChEMBL
Affinity DataIC50: 230nMAssay Description:Inhibition of Burkholderia cenocepacia HldA assessed as ATP depletion after 30 mins by luminescence assayMore data for this Ligand-Target Pair
Affinity DataIC50: 280nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 530nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 650nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
TargetD-beta-D-heptose 7-phosphate kinase(Burkholderia cepacia (strain J2315 / LMG 16656) (B...)
McMaster University
Curated by ChEMBL
McMaster University
Curated by ChEMBL
Affinity DataIC50: 810nMAssay Description:Inhibition of Burkholderia cenocepacia HldA assessed as ATP depletion after 30 mins by luminescence assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 2.20E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 2.70E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 3.30E+3nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 3.40E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 3.70E+3nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 3.80E+3nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 4.30E+3nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 4.50E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 5.60E+3nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 5.90E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 6.90E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 9.00E+3nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 1.26E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 1.30E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 1.42E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+4nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 1.51E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 1.89E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 1.98E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 2.49E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 3.40E+4nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 3.46E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 3.47E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 3.51E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 3.70E+4nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 4.06E+4nMpH: 7.0 T: 2°CAssay Description:The DHFR inhibition observed with each derivative was characterized by determination and comparison of the IC50 values. IC50 determinations were perf...More data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+4nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 8.20E+4nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+5nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+5nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair
Affinity DataIC50: 5.60E+5nMAssay Description:Inhibition of Escherichia coli CBLMore data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)