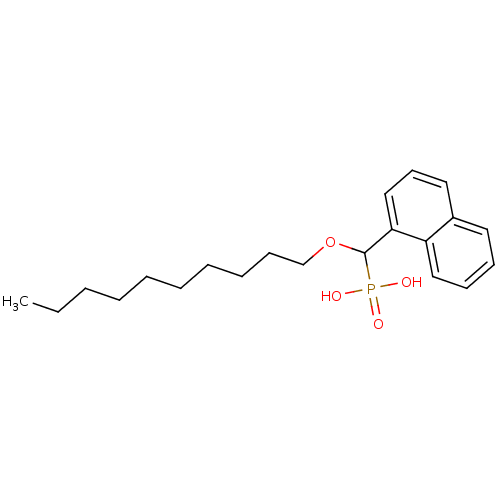
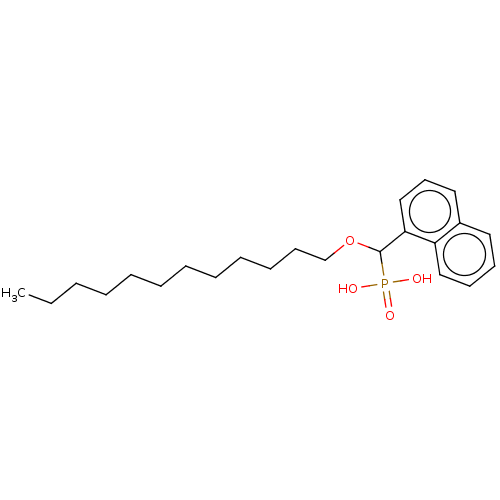
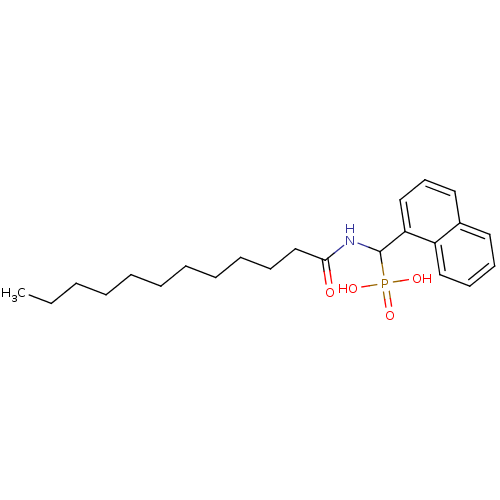
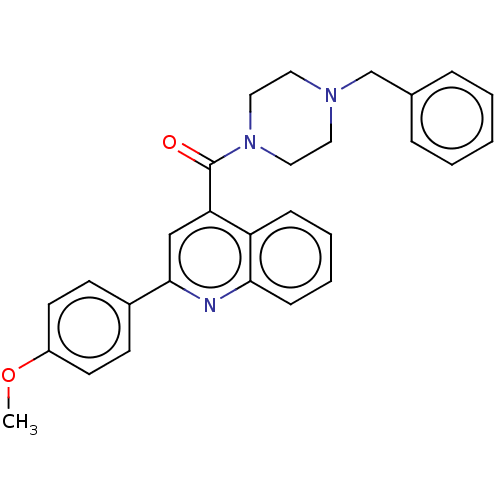
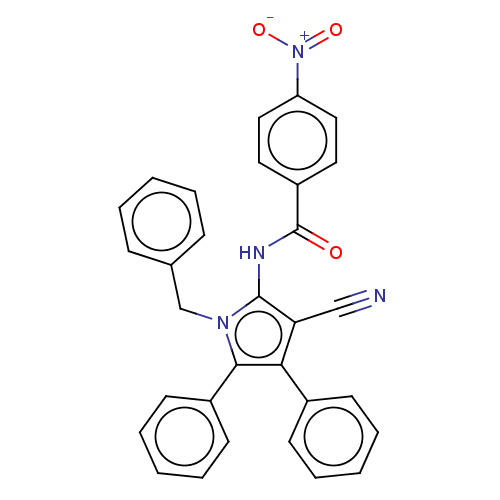
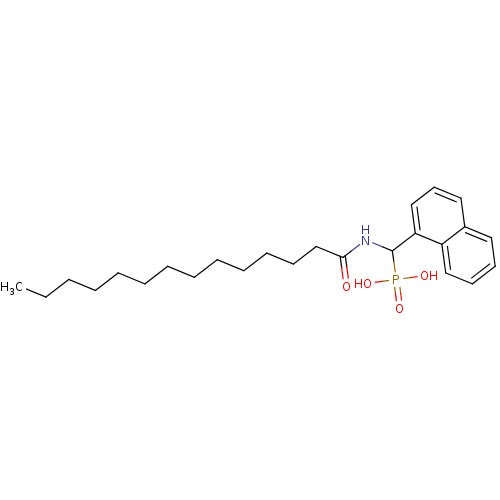
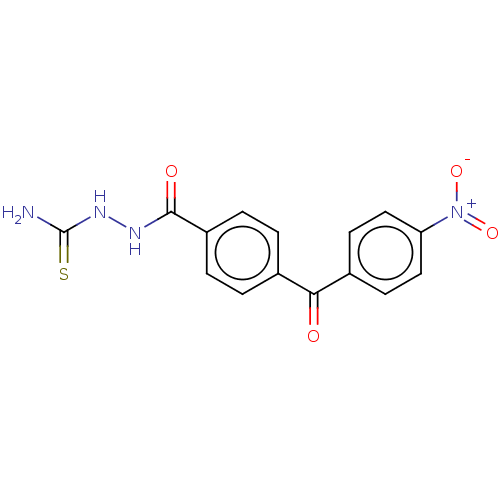
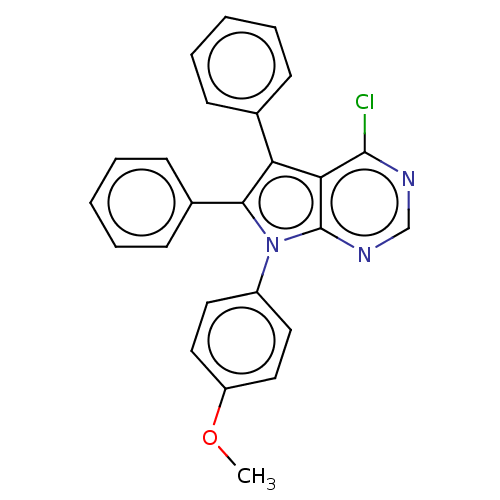
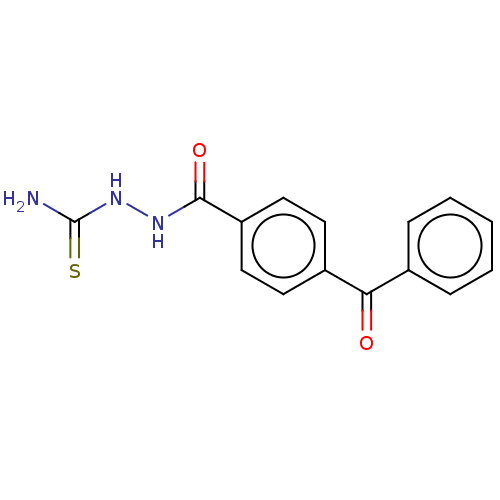
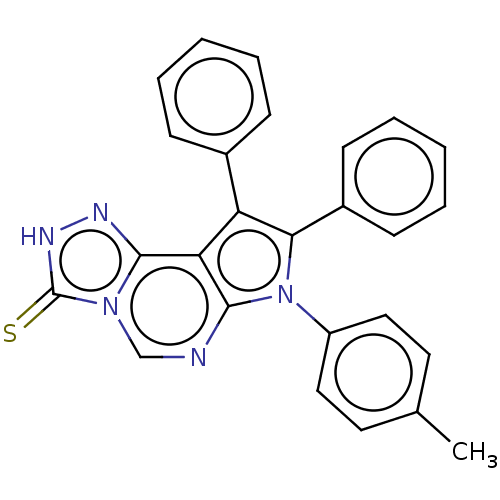
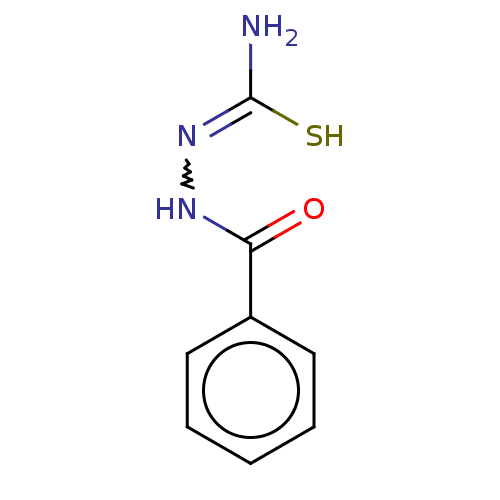
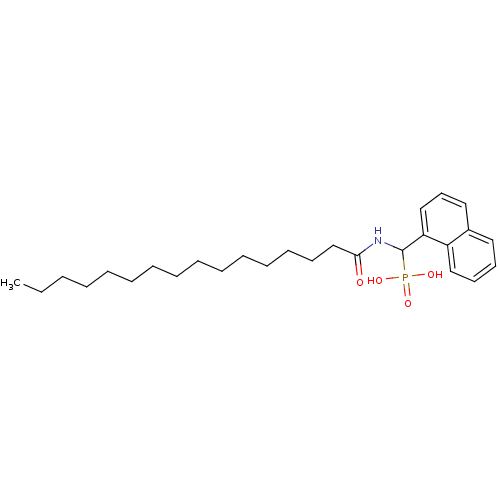
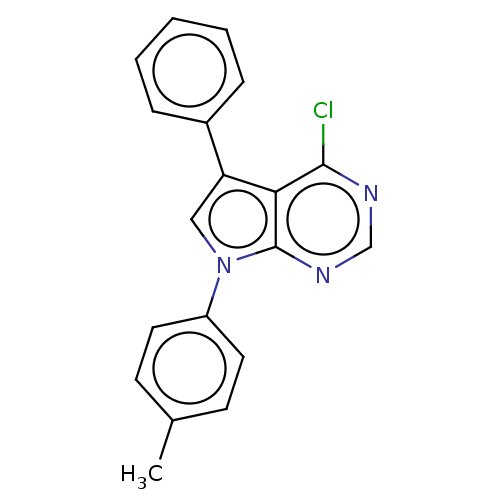
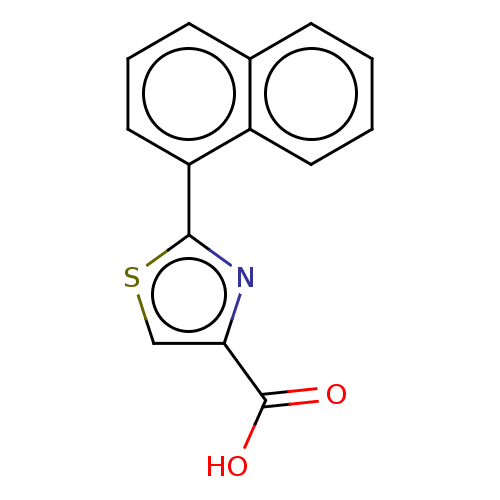
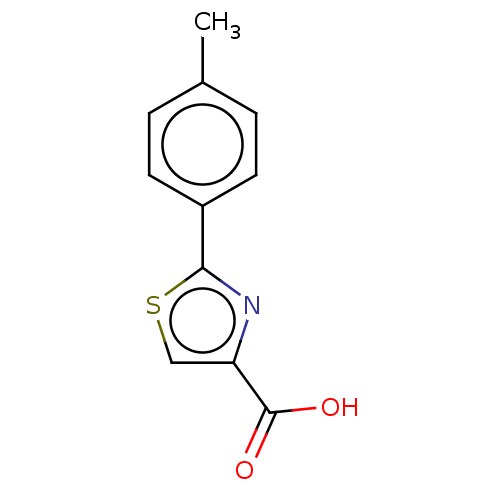
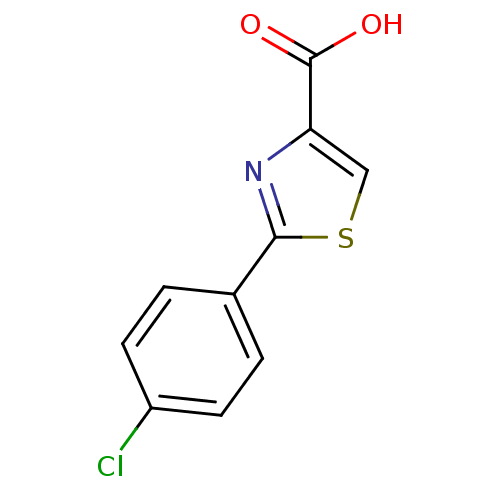
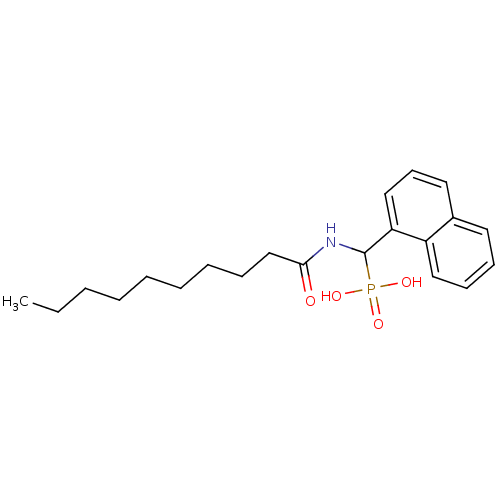
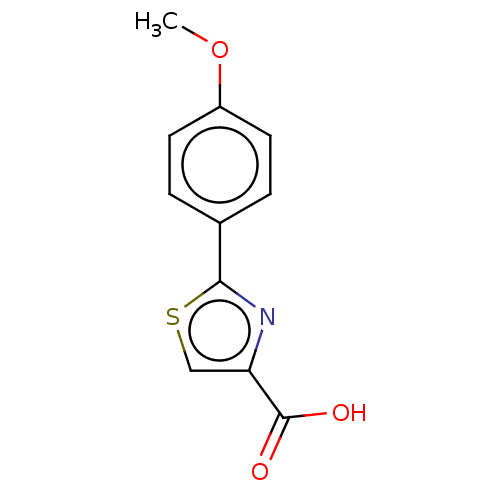
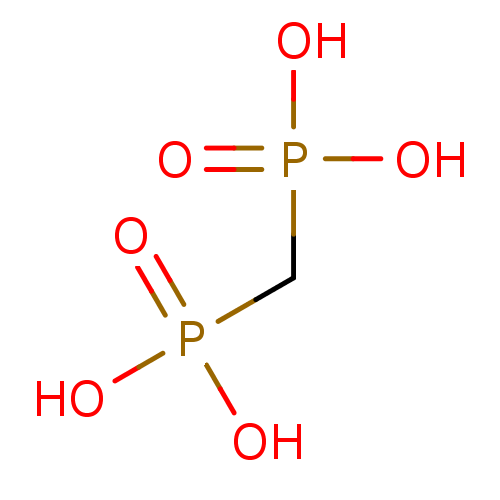
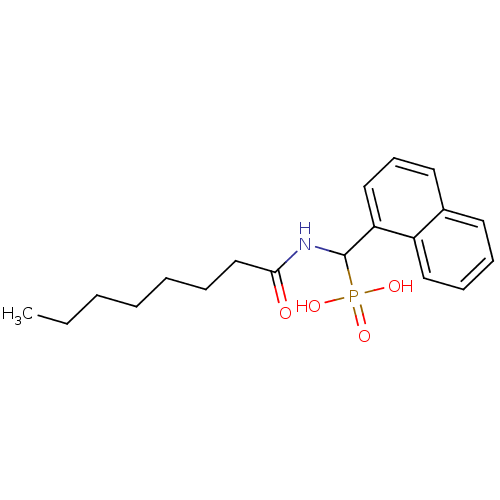
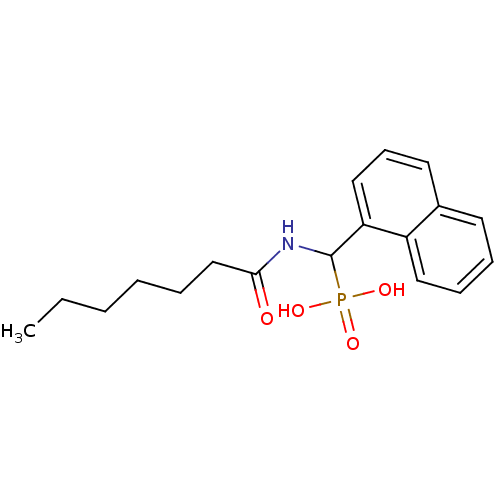
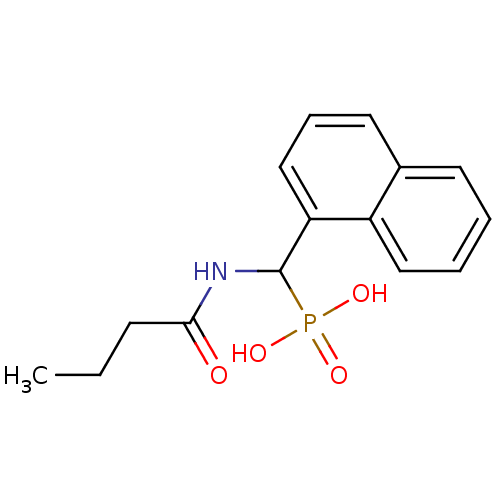
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 200nMAssay Description:Competitive inhibition of red kidney bean PAP using varying levels of pNPP as substrate measured at pH 6.2 by UV-vis spectrophotometric analysisMore data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 400nMAssay Description:Competitive inhibition of red kidney bean PAP using varying levels of pNPP as substrate measured at pH 6.2 by UV-vis spectrophotometric analysisMore data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 500nMAssay Description:Competitive inhibition of red kidney bean PAP using varying levels of pNPP as substrate measured at pH 6.2 by UV-vis spectrophotometric analysisMore data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 800nMAssay Description:Competitive inhibition of red kidney bean PAP using varying levels of pNPP as substrate measured at pH 6.2 by UV-vis spectrophotometric analysisMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase AIM-1(Pseudomonas aeruginosa)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.60E+3nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa recombinant AIM1 expressed in Escherichia coli BL21(DE3) using cefuroxime as substrate by UV/Vis mul...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase AIM-1(Pseudomonas aeruginosa)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 2.20E+3nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa recombinant AIM1 expressed in Escherichia coli BL21(DE3) using cefuroxime as substrate by UV/Vis mul...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 4.00E+3nMpH: 6.2Assay Description:Inhibition of red kidney bean PAP at pH 6.2More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 4.50E+3nMAssay Description:Competitive inhibition of red kidney bean PAP using varying levels of pNPP as substrate measured at pH 6.2 by UV-vis spectrophotometric analysisMore data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 5.00E+3nMpH: 4.9Assay Description:Competitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometryMore data for this Ligand-Target Pair
Affinity DataKi: 5.20E+3nMAssay Description:Mixed inhibition of equine serum BChE using butyrylthiocholine iodide as substrateMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase AIM-1(Pseudomonas aeruginosa)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.09E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa recombinant AIM1 expressed in Escherichia coli BL21(DE3) using cefuroxime as substrate by UV/Vis mul...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.10E+4nMpH: 4.9Assay Description:Competitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometryMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.10E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa beta-lactamase IMP-1 assessed as formation of 4-nitrothiophenolate at pH 7 using CENTA as chromogeni...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Aeromonas hydrophila)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.10E+4nMAssay Description:Competitive inhibition of Aeromonas hydrophila recombinant CphA expressed in Escherichia coli BL21(DE3) using meropenem as substrate by UV/Vis multi-...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.20E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa metallo-beta-lactamase IMP-1 expressed in Escherichia coli BL21(DE3) using CENTA as substrate by spe...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.30E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa beta-lactamase IMP-1 assessed as formation of 4-nitrothiophenolate at pH 7 using CENTA as chromogeni...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.40E+4nMAssay Description:Competitive inhibition of red kidney bean PAP using varying levels of pNPP as substrate measured at pH 6.2 by UV-vis spectrophotometric analysisMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.40E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa beta-lactamase IMP-1 assessed as formation of 4-nitrothiophenolate at pH 7 using CENTA as chromogeni...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.50E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa metallo-beta-lactamase IMP-1 expressed in Escherichia coli BL21(DE3) using CENTA as substrate by spe...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.60E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa beta-lactamase IMP-1 assessed as formation of 4-nitrothiophenolate at pH 7 using CENTA as chromogeni...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.80E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa metallo-beta-lactamase IMP-1 expressed in Escherichia coli BL21(DE3) using CENTA as substrate by spe...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.80E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa beta-lactamase IMP-1 assessed as formation of 4-nitrothiophenolate at pH 7 using CENTA as chromogeni...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.90E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa beta-lactamase IMP-1 assessed as formation of 4-nitrothiophenolate at pH 7 using CENTA as chromogeni...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.90E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa metallo-beta-lactamase IMP-1 expressed in Escherichia coli BL21(DE3) using CENTA as substrate by spe...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 2.90E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa metallo-beta-lactamase IMP-1 expressed in Escherichia coli BL21(DE3) using CENTA as substrate by spe...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 3.10E+4nMpH: 4.9Assay Description:Competitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometryMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 3.30E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa metallo-beta-lactamase IMP-1 expressed in Escherichia coli BL21(DE3) using CENTA as substrate by spe...More data for this Ligand-Target Pair
TargetTartrate-resistant acid phosphatase type 5(Sus scrofa)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 3.30E+4nMAssay Description:Competitive inhibition of pig purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV/Vis spectro...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 4.10E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa beta-lactamase IMP-1 assessed as formation of 4-nitrothiophenolate at pH 7 using CENTA as chromogeni...More data for this Ligand-Target Pair
TargetTartrate-resistant acid phosphatase type 5(Sus scrofa)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 4.20E+4nMAssay Description:Competitive inhibition of pig purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV/Vis spectro...More data for this Ligand-Target Pair
TargetTartrate-resistant acid phosphatase type 5(Sus scrofa)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 4.90E+4nMAssay Description:Competitive inhibition of pig purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV/Vis spectro...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 5.00E+4nMpH: 6.2Assay Description:Inhibition of red kidney bean PAP at pH 6.2More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Aeromonas hydrophila)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 5.20E+4nMAssay Description:Competitive inhibition of Aeromonas hydrophila recombinant CphA expressed in Escherichia coli BL21(DE3) using meropenem as substrate by UV/Vis multi-...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 5.70E+4nMpH: 4.9Assay Description:Competitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometryMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 7.50E+4nMAssay Description:Competitive inhibition of Pseudomonas aeruginosa beta-lactamase IMP-1 assessed as formation of 4-nitrothiophenolate at pH 7 using CENTA as chromogeni...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.02E+5nMpH: 4.9Assay Description:Uncompetitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometr...More data for this Ligand-Target Pair
TargetTartrate-resistant acid phosphatase type 5(Sus scrofa)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.20E+5nMAssay Description:Competitive inhibition of pig purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV/Vis spectro...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.40E+5nMAssay Description:Inhibition of red kidney beans purple acid phosphataseMore data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.60E+5nMAssay Description:Inhibition of red kidney beans purple acid phosphataseMore data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.60E+5nMAssay Description:Inhibition of red kidney bean PAP using pNPP as substrateMore data for this Ligand-Target Pair
Affinity DataKi: 1.85E+5nMAssay Description:Competitive inhibition of red kidney bean purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV...More data for this Ligand-Target Pair
TargetTartrate-resistant acid phosphatase type 5(Sus scrofa)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.90E+5nMAssay Description:Competitive inhibition of pig purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV/Vis spectro...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.95E+5nMpH: 4.9Assay Description:Competitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometryMore data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 2.22E+5nMpH: 4.9Assay Description:Competitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometryMore data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 2.38E+5nMpH: 4.9Assay Description:Competitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometryMore data for this Ligand-Target Pair
Affinity DataKi: 3.40E+5nMAssay Description:Competitive inhibition of red kidney bean purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV...More data for this Ligand-Target Pair
Affinity DataKi: 4.00E+5nMAssay Description:Competitive inhibition of red kidney bean purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 4.43E+5nMpH: 4.9Assay Description:Uncompetitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometr...More data for this Ligand-Target Pair
TargetFe(3+)-Zn(2+) purple acid phosphatase(Phaseolus vulgaris)
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 4.46E+5nMpH: 4.9Assay Description:Uncompetitive inhibition of red kidney bean PAP using para-nitrophenol as substrate at pH 4.9 preincubated for 10 mins by UV-visible spectrophotometr...More data for this Ligand-Target Pair
Affinity DataKi: 5.30E+5nMAssay Description:Competitive inhibition of red kidney bean purple acid phosphatase Fe(3)Fe(2) state assessed as enzyme-inhibitor complex using pNPP as substrate by UV...More data for this Ligand-Target Pair



 3D Structure (crystal)
3D Structure (crystal)