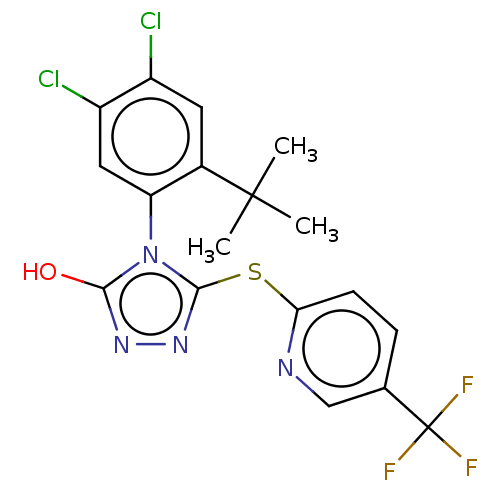
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 1nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair
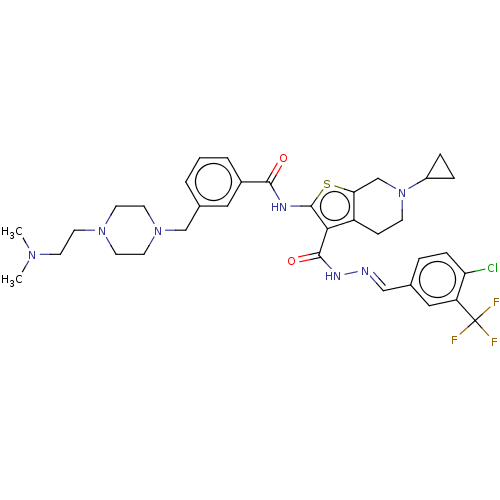
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 1.70nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair
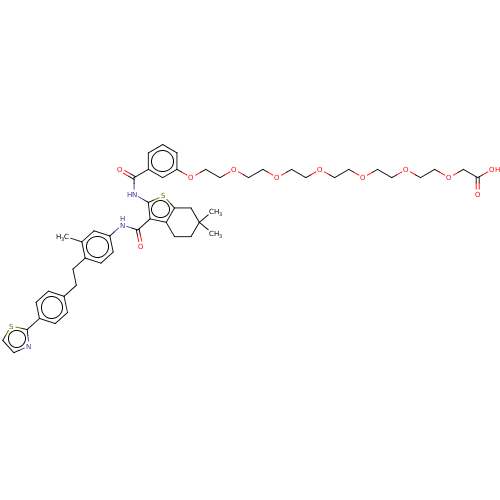
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 1.40nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair
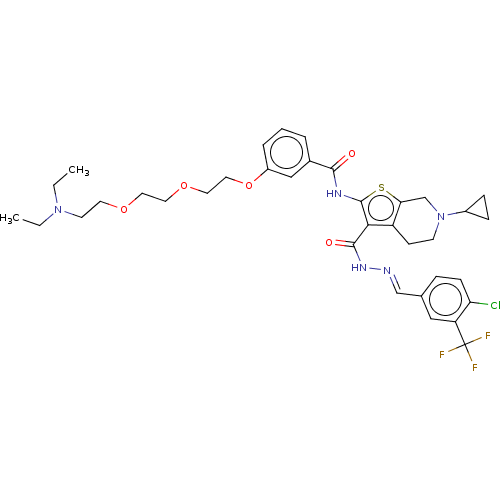
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 5.30nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 3nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 32nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 7nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 7.30nMAssay Description:Transactivation of chimeric GAL4-fused human PPARgamma transfected in HEK293EBNA cells measured at 24 hrs after drug treatment by steady-Glo lucifera...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 5.30nMAssay Description:Transactivation of chimeric GAL4-fused human PPARgamma transfected in HEK293EBNA cells measured at 24 hrs after drug treatment by steady-Glo lucifera...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 13nMAssay Description:Transactivation of chimeric GAL4-fused human PPARgamma transfected in HEK293EBNA cells measured at 24 hrs after drug treatment by steady-Glo lucifera...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 3.60nMAssay Description:Transactivation of chimeric GAL4-fused human PPARgamma transfected in HEK293EBNA cells measured at 24 hrs after drug treatment by steady-Glo lucifera...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 21nMAssay Description:Transactivation of chimeric GAL4-fused human PPARgamma transfected in HEK293EBNA cells measured at 24 hrs after drug treatment by steady-Glo lucifera...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 24nMAssay Description:Transactivation of chimeric GAL4-fused human PPARgamma transfected in HEK293EBNA cells measured at 24 hrs after drug treatment by steady-Glo lucifera...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 4.40nMAssay Description:Transactivation of chimeric GAL4-fused human PPARgamma transfected in HEK293EBNA cells measured at 24 hrs after drug treatment by steady-Glo lucifera...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 12nMAssay Description:Transactivation of chimeric GAL4-fused human PPARgamma transfected in HEK293EBNA cells measured at 24 hrs after drug treatment by steady-Glo lucifera...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 3.30nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Homo sapiens (Human))
Hokkaido University
Curated by ChEMBL
Hokkaido University
Curated by ChEMBL
Affinity DataEC50: 19nMAssay Description:Agonist activity at PPARgamma in human MKN45 cells assessed as induction of cell aggregation incubated for 5 days by Hoechst 33342 staining based IN ...More data for this Ligand-Target Pair

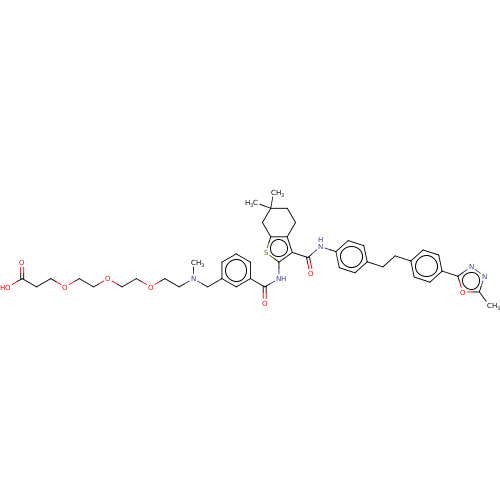
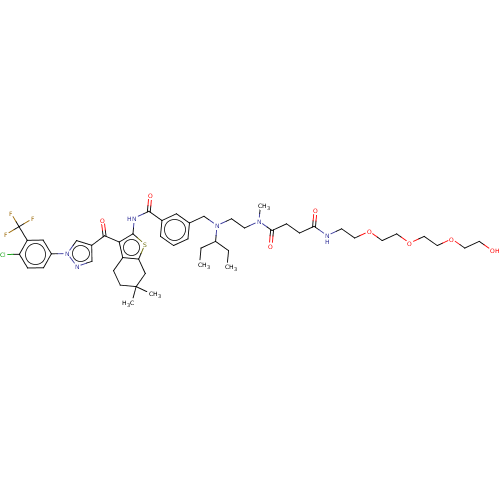
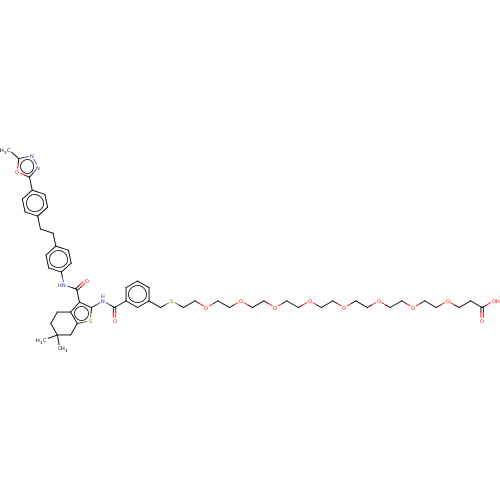
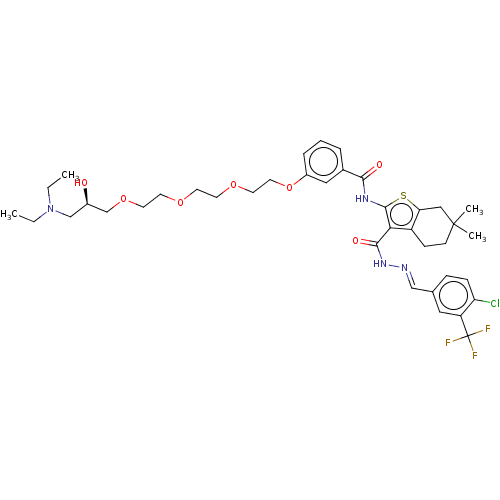
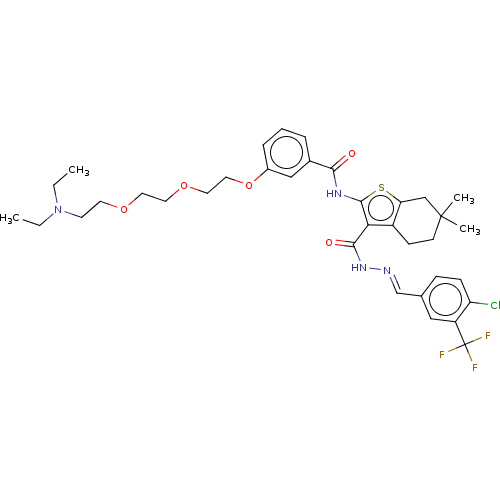

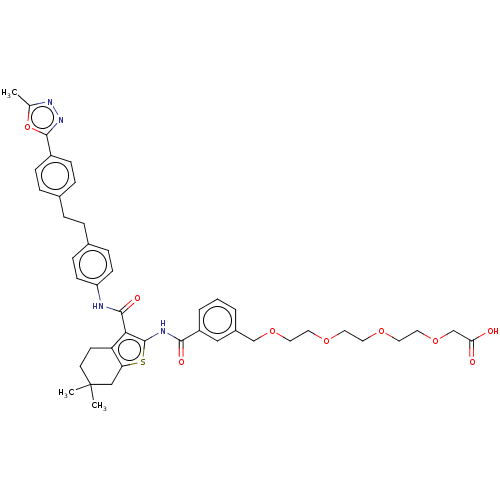
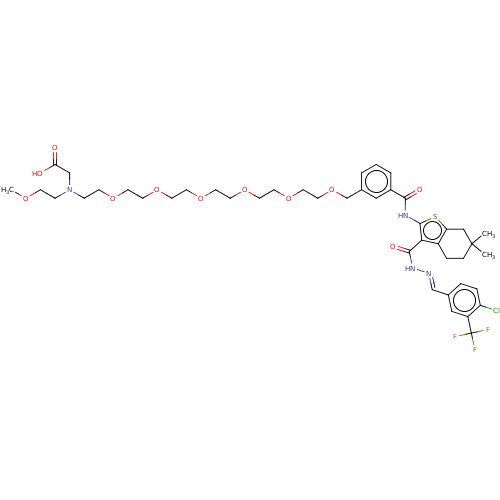
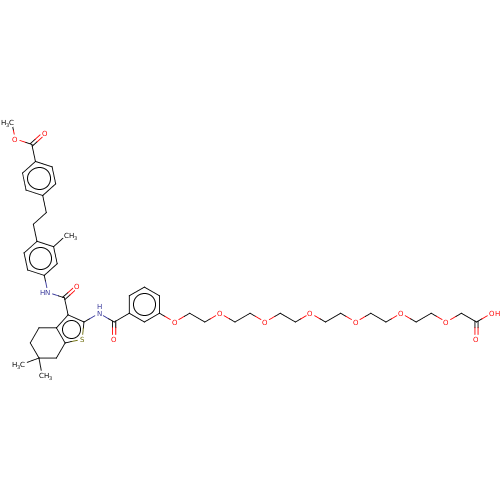

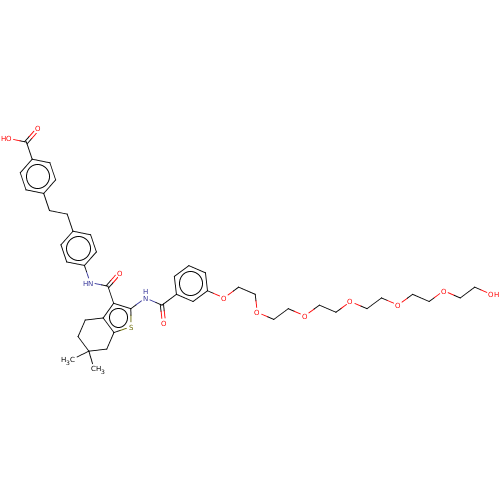
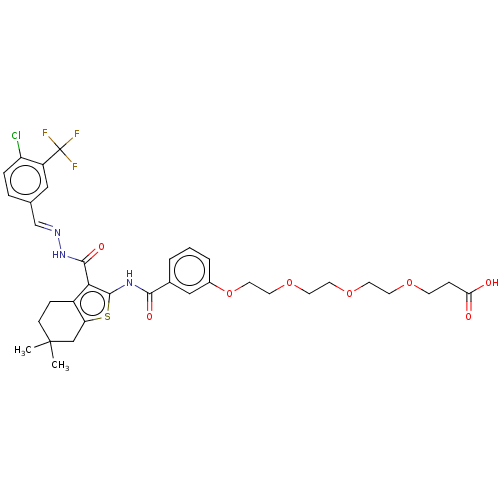
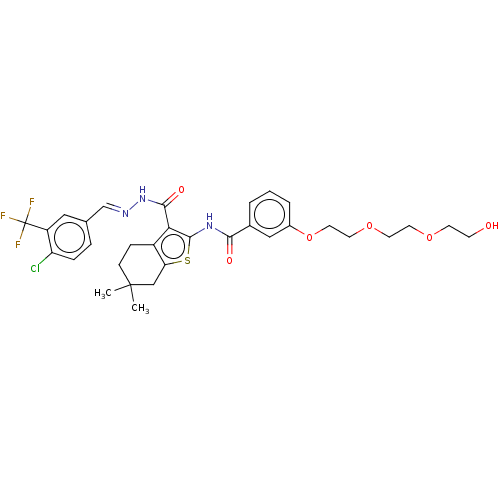
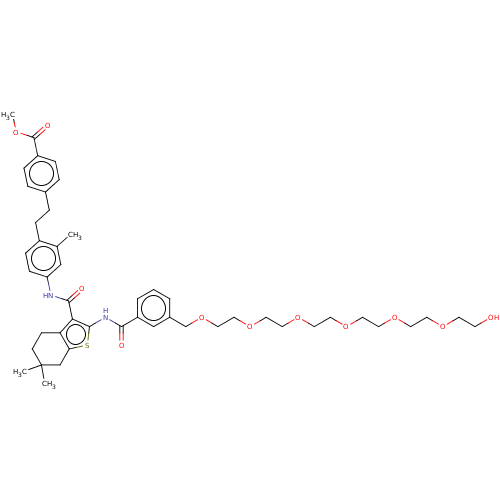

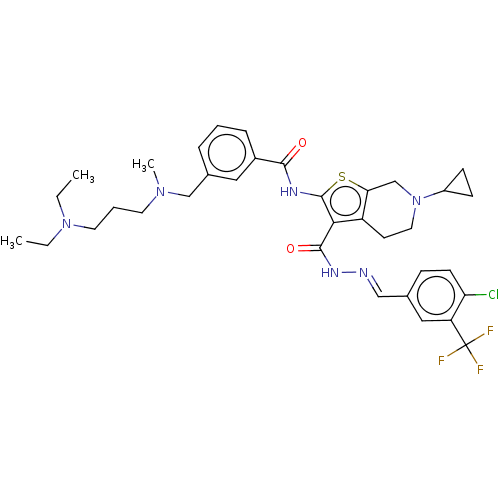
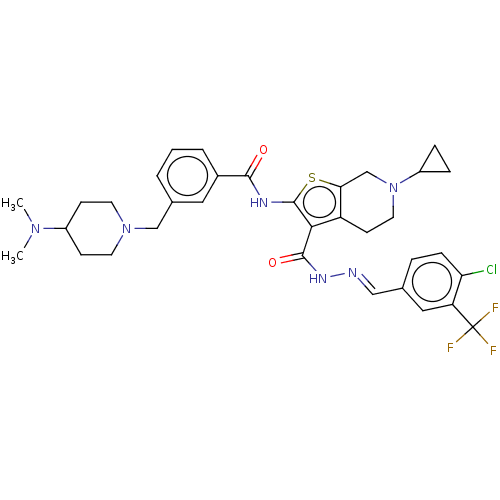
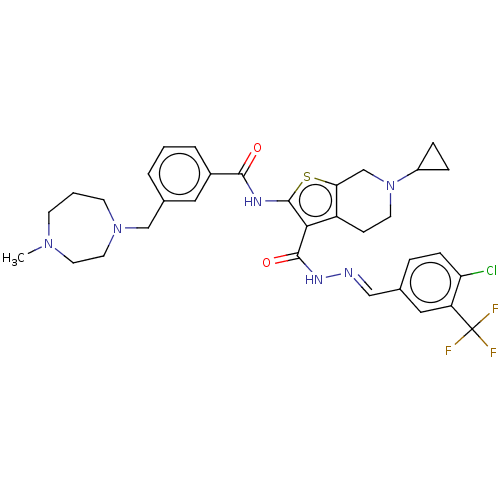
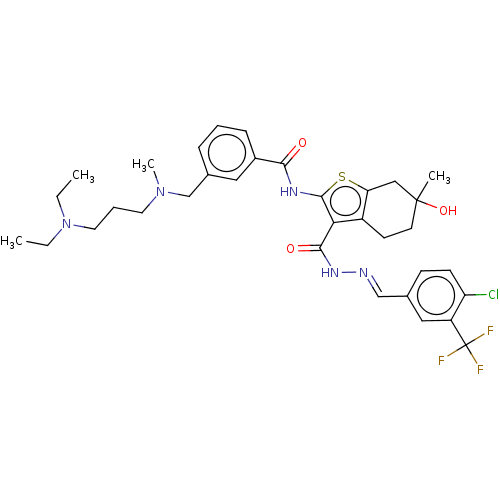
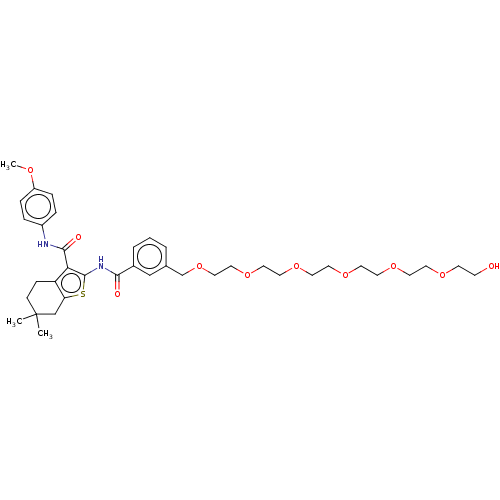
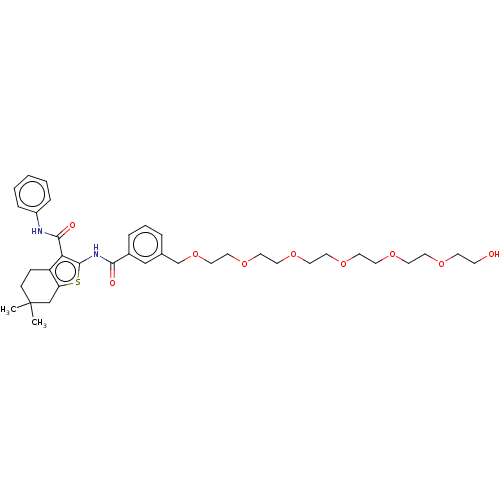
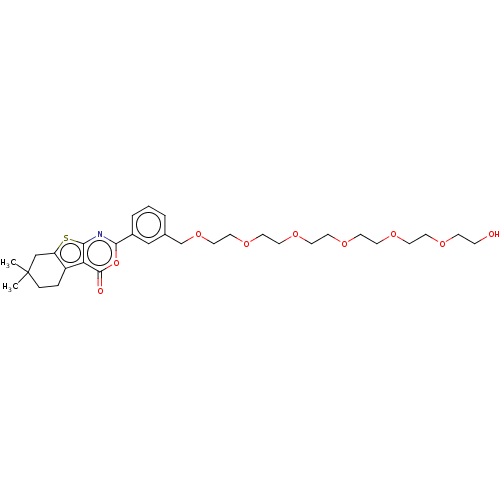
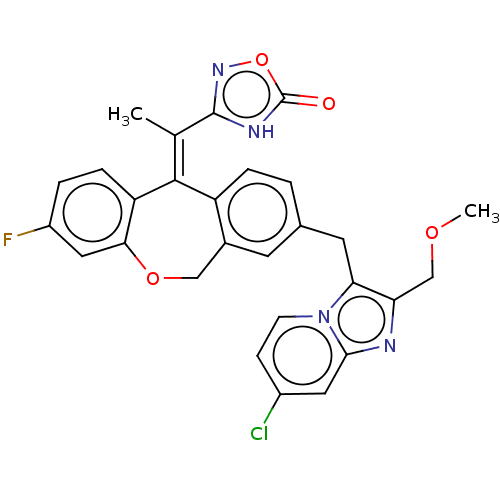
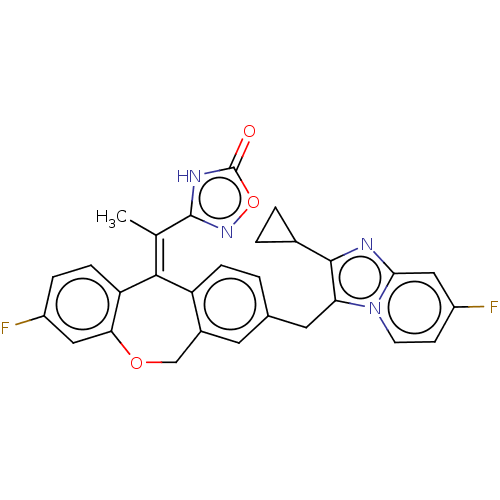
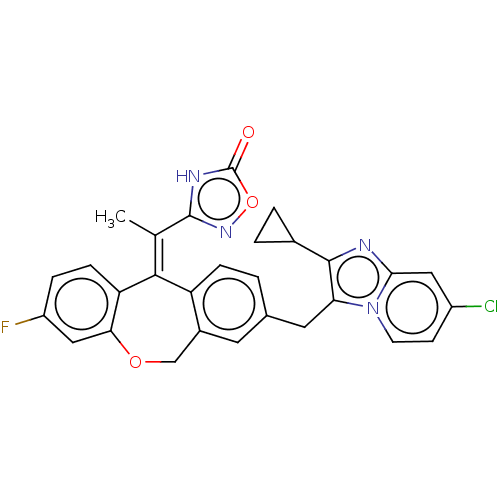
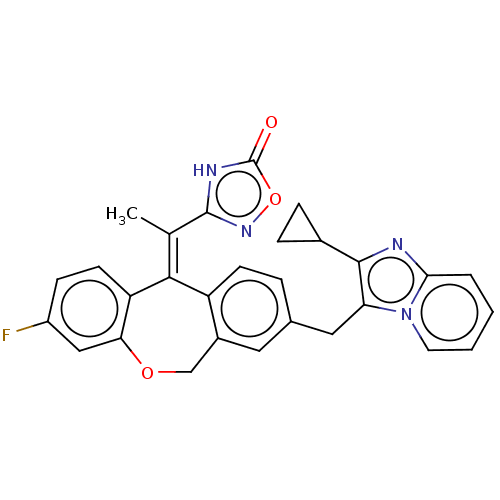
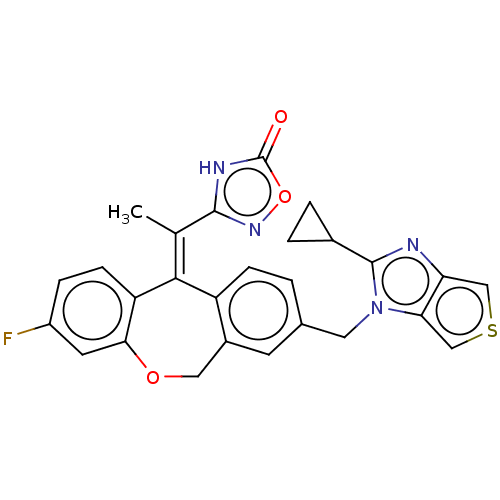
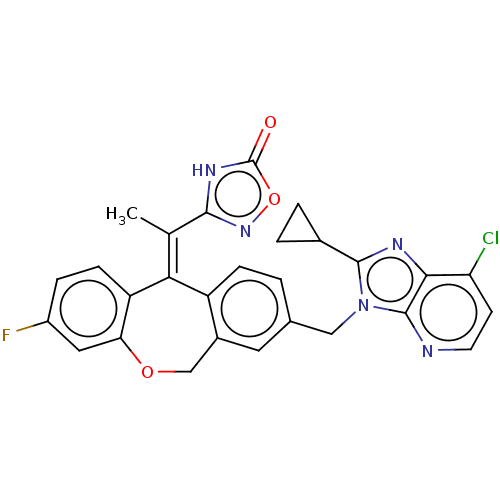
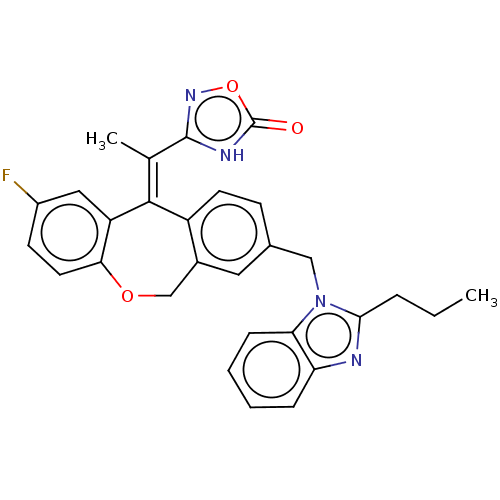
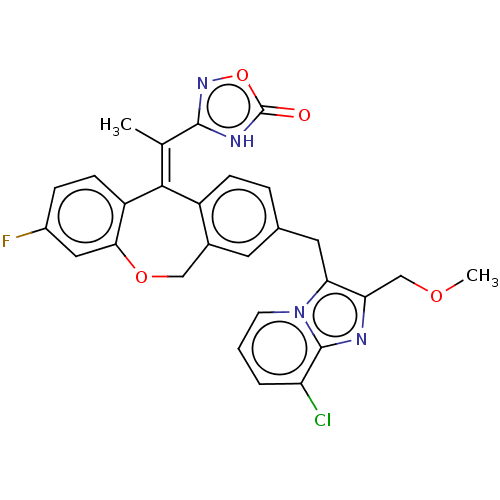

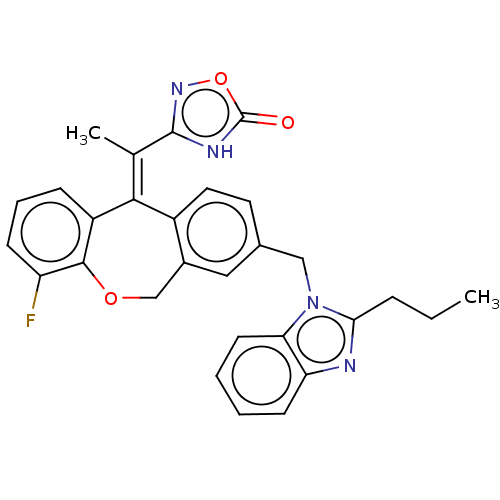
 3D Structure (crystal)
3D Structure (crystal)