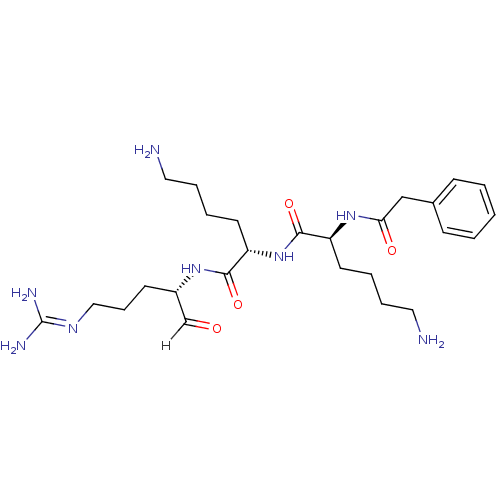
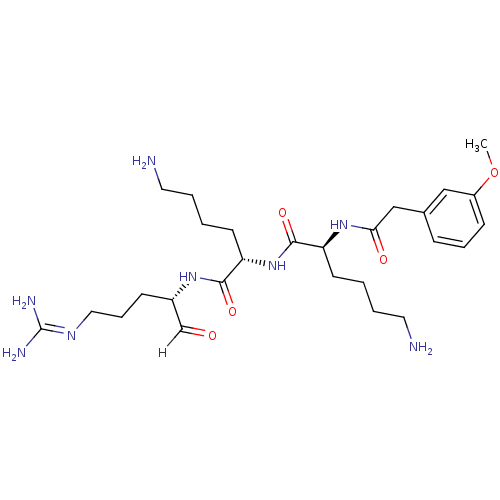
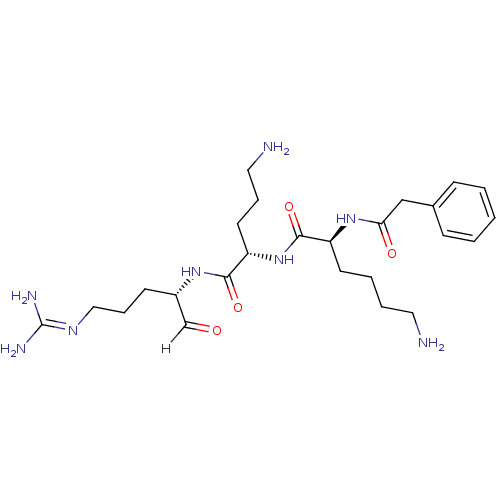
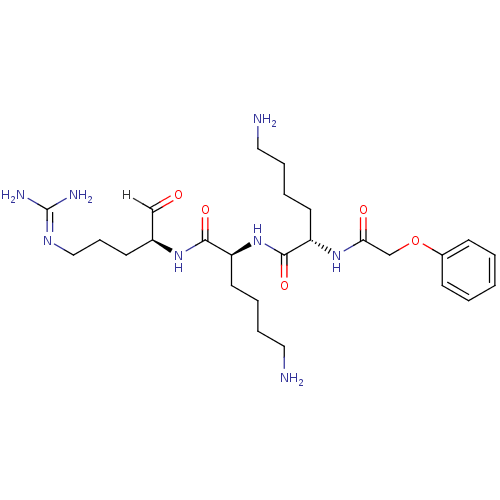
Affinity DataKi: 0.400nMAssay Description:Inhibition of factor-10a (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 1nMAssay Description:Inhibition of human coagulation factor 10a using S-2765 as substrate measured up to 20 mins by chromogenic assayMore data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 3.5nMAssay Description:Inhibition of recombinant human N-terminal His-tagged SIRT5 (34 to 269 residues) expressed in Escherichia coli Transetta(DE3) cells using Benzyl Lys(...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 3.5nMAssay Description:Inhibition of recombinant human N-terminal His-tagged SIRT5 (34 to 269 residues) expressed in Escherichia coli Transetta(DE3) cells using Benzyl Lys(...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 3.5nMAssay Description:Inhibition of recombinant human N-terminal His-tagged SIRT5 (34 to 269 residues) expressed in Escherichia coli Transetta(DE3) cells using Benzyl Lys(...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 3.5nMAssay Description:Inhibition of human recombinant N-terminal Strep2-tagged SIRT5 expressed in Escherichia coli BL21 (DE3) using Abz-GVLK(glutaryl)AY(NO2)GV-NH2 as subs...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 3.5nMAssay Description:Inhibition of human recombinant N-terminal Strep2-tagged SIRT5 expressed in Escherichia coli BL21 (DE3) using Abz-GVLK(glutaryl)AY(NO2)GV-NH2 as subs...More data for this Ligand-Target Pair
Affinity DataKi: 6nM ΔG°: -48.8kJ/mole IC50: 32nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 7nMAssay Description:Inhibition of human recombinant N-terminal Strep2-tagged SIRT5 expressed in Escherichia coli BL21 (DE3) using Abz-GVLK(glutaryl)AY(NO2)GV-NH2 as subs...More data for this Ligand-Target Pair
Affinity DataKi: 8nM ΔG°: -47.0kJ/mole IC50: 50nMpH: 7.0 T: 2°CAssay Description:Activity was measured as a change in absorbance over time at a wavelength of 340 nm (A340), so as to monitor the disappearance of NADPH. After incuba...More data for this Ligand-Target Pair
Affinity DataKi: 9nM ΔG°: -47.8kJ/mole IC50: 51nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 9nM ΔG°: -46.7kJ/mole IC50: 90nMpH: 7.0 T: 2°CAssay Description:Activity was measured as a change in absorbance over time at a wavelength of 340 nm (A340), so as to monitor the disappearance of NADPH. After incuba...More data for this Ligand-Target Pair
Affinity DataKi: 11nM ΔG°: -47.3kJ/mole IC50: 60nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 13nM ΔG°: -46.8kJ/mole IC50: 73nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 14nMAssay Description:Competitive inhibition of human recombinant N-terminal Strep2-tagged SIRT5 expressed in Escherichia coli BL21 (DE3) using Abz-GVLK(glutaryl)AY(NO2)GV...More data for this Ligand-Target Pair
Affinity DataKi: 19nM ΔG°: -45.8kJ/mole IC50: 107nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 20nM ΔG°: -45.7kJ/mole IC50: 112nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 23nMAssay Description:Inhibition of recombinant human N-terminal His-tagged SIRT5 (34 to 269 residues) expressed in Escherichia coli Transetta(DE3) cells using Benzyl Lys(...More data for this Ligand-Target Pair
Affinity DataKi: 26nM ΔG°: -45.0kJ/mole IC50: 146nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 27nM ΔG°: -43.9kJ/mole IC50: 50nMpH: 7.0 T: 2°CAssay Description:Activity was measured as a change in absorbance over time at a wavelength of 340 nm (A340), so as to monitor the disappearance of NADPH. After incuba...More data for this Ligand-Target Pair
Affinity DataKi: 28nM ΔG°: -44.8kJ/mole IC50: 154nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 40nM ΔG°: -43.9kJ/mole IC50: 222nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 40nM ΔG°: -42.9kJ/mole IC50: 600nMpH: 7.0 T: 2°CAssay Description:Activity was measured as a change in absorbance over time at a wavelength of 340 nm (A340), so as to monitor the disappearance of NADPH. After incuba...More data for this Ligand-Target Pair
Affinity DataKi: 41nM ΔG°: -43.9kJ/mole IC50: 231nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 44nM ΔG°: -43.7kJ/mole IC50: 245nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 46nM ΔG°: -43.6kJ/mole IC50: 255nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 49nM ΔG°: -43.4kJ/mole IC50: 271nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 53nM ΔG°: -43.2kJ/mole IC50: 297nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 58nM ΔG°: -43.0kJ/mole IC50: 325nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 81nM ΔG°: -42.1kJ/mole IC50: 454nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 83nMAssay Description:Competitive inhibition of human recombinant N-terminal Strep2-tagged SIRT5 expressed in Escherichia coli BL21 (DE3) using Abz-GVLK(glutaryl)AY(NO2)GV...More data for this Ligand-Target Pair
Affinity DataKi: 90nM ΔG°: -40.9kJ/mole IC50: 900nMpH: 7.0 T: 2°CAssay Description:Activity was measured as a change in absorbance over time at a wavelength of 340 nm (A340), so as to monitor the disappearance of NADPH. After incuba...More data for this Ligand-Target Pair
Affinity DataKi: 104nM ΔG°: -41.5kJ/mole IC50: 580nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 111nM ΔG°: -41.3kJ/mole IC50: 619nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 135nMAssay Description:Inhibition of human recombinant N-terminal Strep2-tagged SIRT5 expressed in Escherichia coli BL21 (DE3) using Abz-GVLK(glutaryl)AY(NO2)GV-NH2 as subs...More data for this Ligand-Target Pair
Affinity DataKi: 160nM ΔG°: -40.3kJ/mole IC50: 891nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 240nM ΔG°: -38.4kJ/mole IC50: 3.00E+3nMpH: 7.0 T: 2°CAssay Description:Activity was measured as a change in absorbance over time at a wavelength of 340 nm (A340), so as to monitor the disappearance of NADPH. After incuba...More data for this Ligand-Target Pair
TargetNAD-dependent protein deacylase sirtuin-5, mitochondrial(Homo sapiens (Human))
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Department of Enzymology, Institute of Biochemistry and Biotechnology , Martin-Luther-University Halle-Wittenberg , 06120 Halle/Saale , Germany.
Curated by ChEMBL
Affinity DataKi: 263nMAssay Description:Inhibition of human recombinant N-terminal Strep2-tagged SIRT5 expressed in Escherichia coli BL21 (DE3) using Abz-GVLK(glutaryl)AY(NO2)GV-NH2 as subs...More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Inhibition of factor-10a (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 2.07E+3nM ΔG°: -33.7kJ/mole IC50: 1.16E+4nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 3.60E+3nMAssay Description:Inhibition of t-PA (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 4.60E+3nM ΔG°: -31.7kJ/mole IC50: 2.57E+4nMpH: 9.5 T: 2°CAssay Description:Inhibition of WNV protease activity was measured using recombinant WNV protease, which first incubated with inhibitor in 96-well plates. Catalysis wa...More data for this Ligand-Target Pair
Affinity DataKi: 5.50E+3nM ΔG°: -30.5kJ/mole IC50: 4.50E+4nMpH: 7.0 T: 2°CAssay Description:Activity was measured as a change in absorbance over time at a wavelength of 340 nm (A340), so as to monitor the disappearance of NADPH. After incuba...More data for this Ligand-Target Pair
Affinity DataKi: 5.60E+3nM ΔG°: -30.5kJ/mole IC50: 4.20E+4nMpH: 7.0 T: 2°CAssay Description:Activity was measured as a change in absorbance over time at a wavelength of 340 nm (A340), so as to monitor the disappearance of NADPH. After incuba...More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nMAssay Description:Inhibition of factor-10a (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 2.90E+4nMAssay Description:Inhibition of plasmin (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMAssay Description:Inhibition of human coagulation factor 10a using S-2765 as substrate measured up to 20 mins by chromogenic assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of human coagulation factor 10a using S-2765 as substrate measured up to 20 mins by chromogenic assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of human coagulation factor 10a using S-2765 as substrate measured up to 20 mins by chromogenic assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.600nMAssay Description:Inhibition of human coagulation factor 10a using S-2765 as substrate measured up to 20 mins by chromogenic assayMore data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)