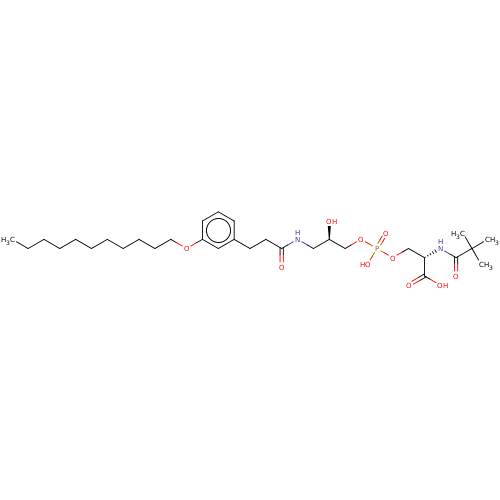
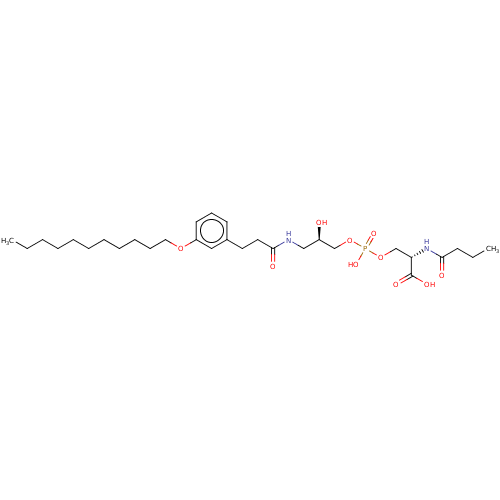
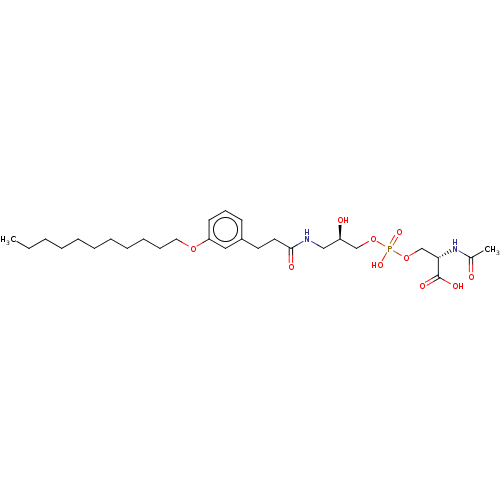
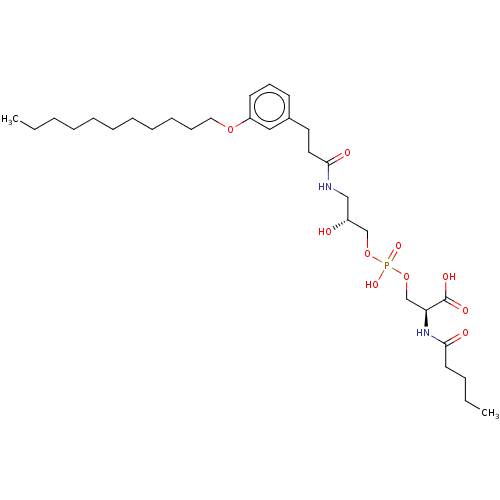
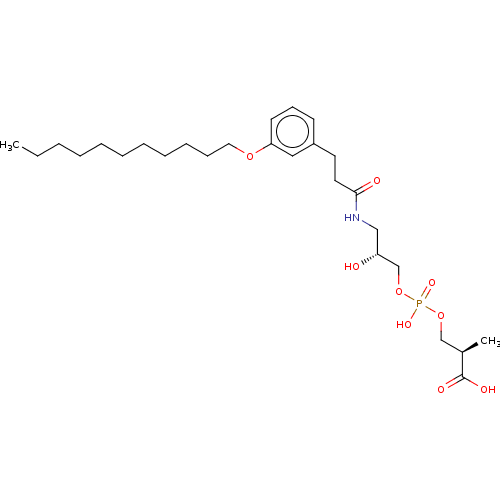
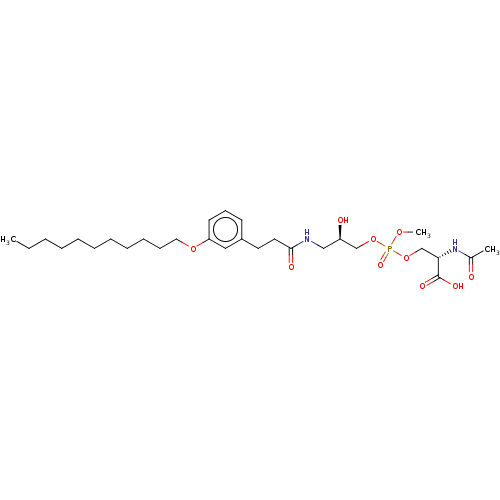
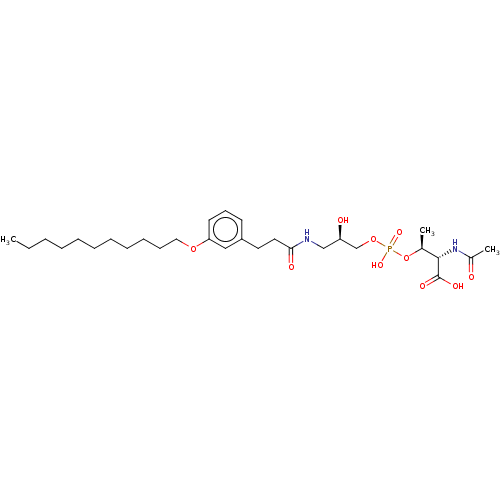
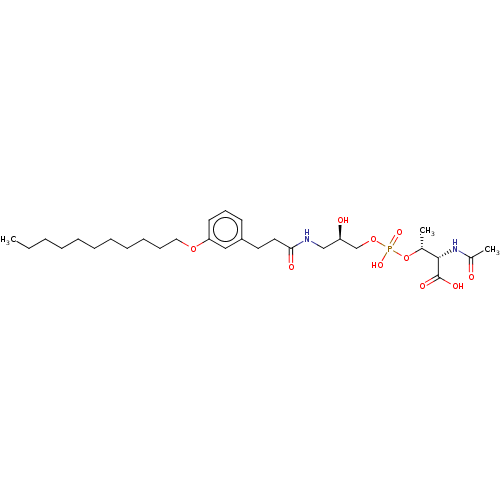
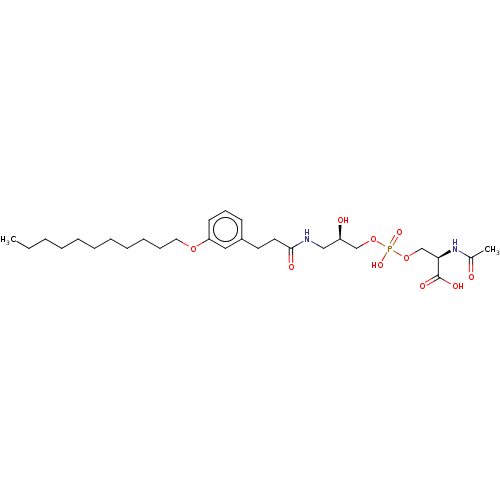
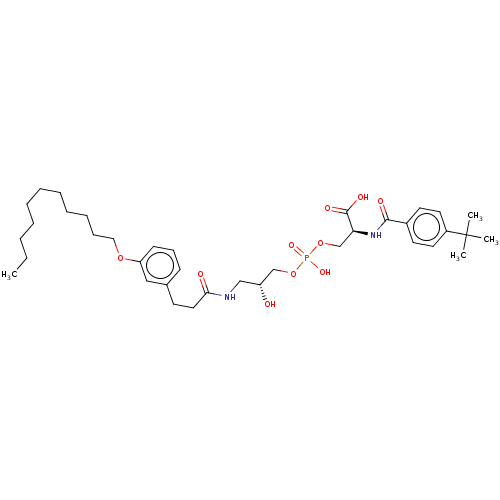
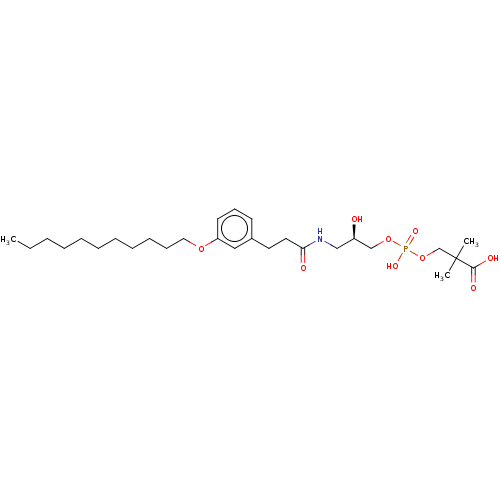
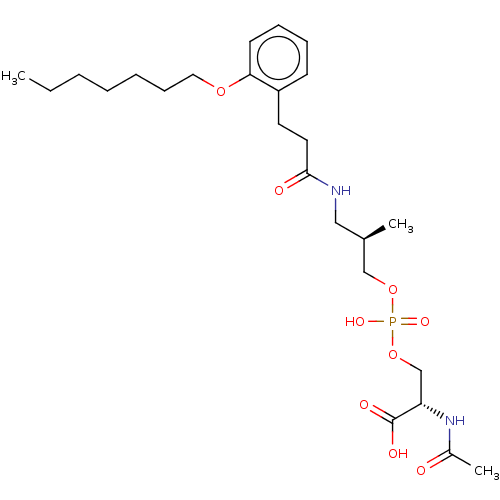
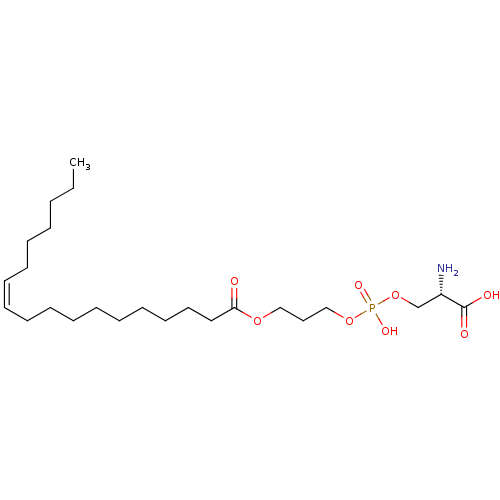
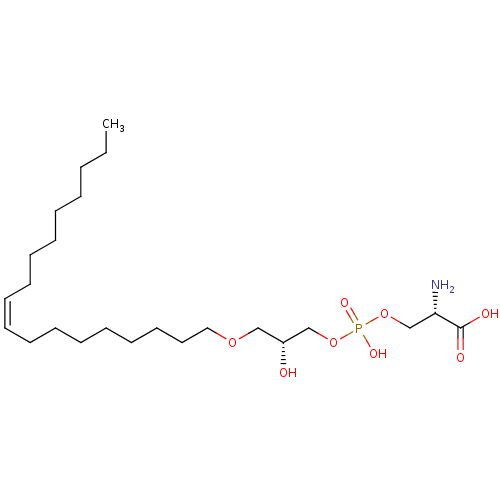
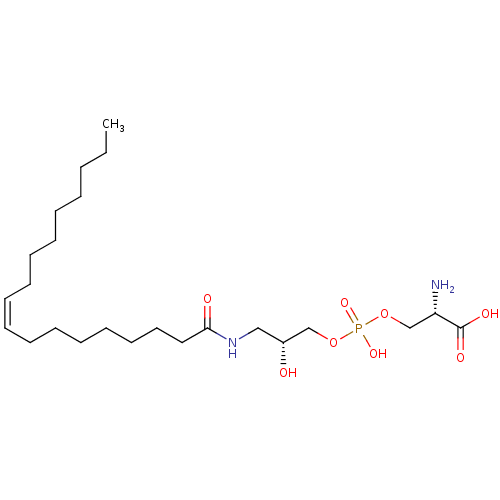
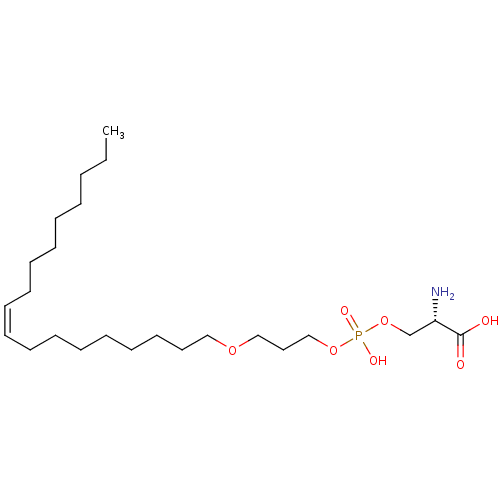
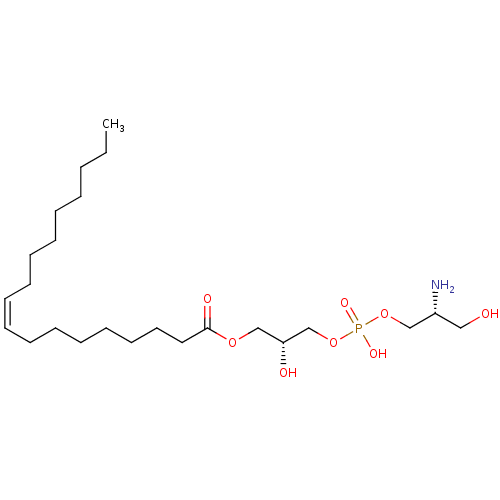
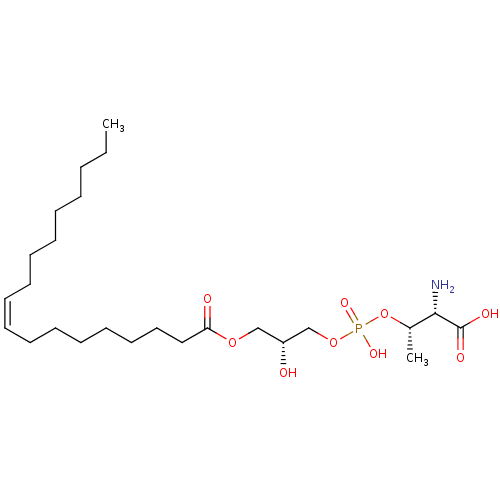
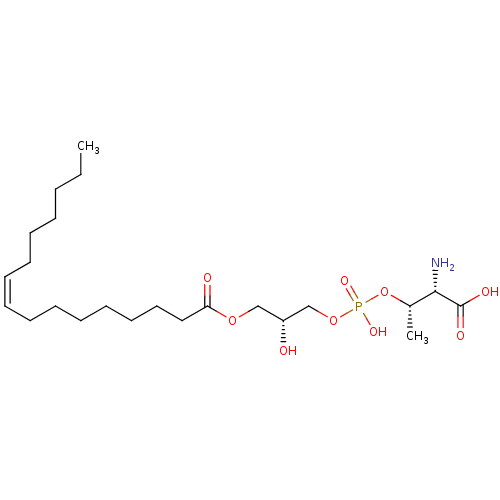
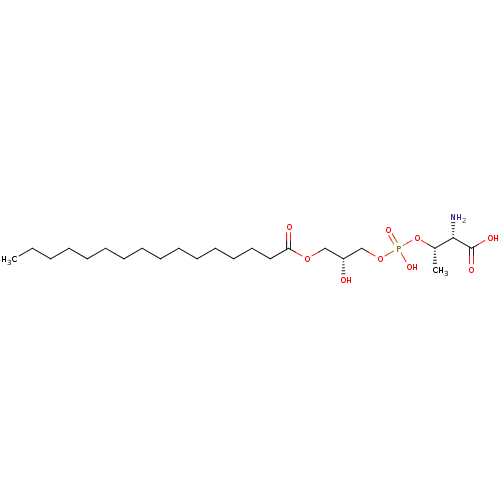
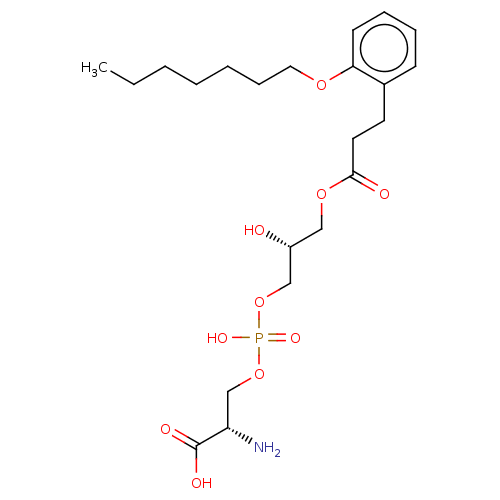
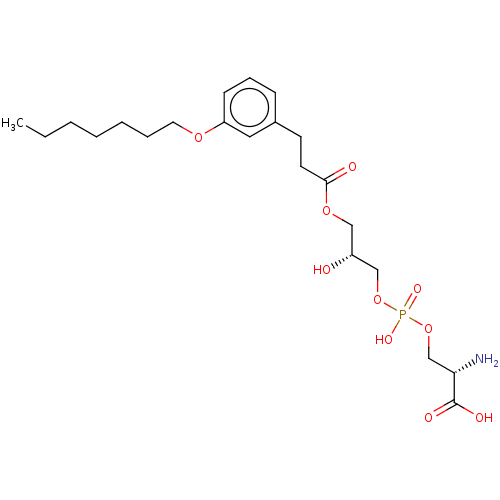
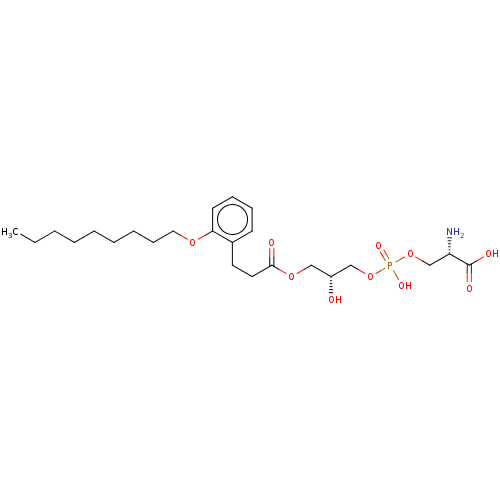
Affinity DataEC50: 170nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 4.50E+3nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 1.20E+3nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 330nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 140nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+3nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+3nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 2.20E+3nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+3nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 2.10E+3nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 210nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 710nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 120nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 100nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 460nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 3.20nMAssay Description:Agonist activity at P2Y10 (unknown origin) transfected in HEK293A cells after 1 hr by TGFalpha shedding assayMore data for this Ligand-Target Pair