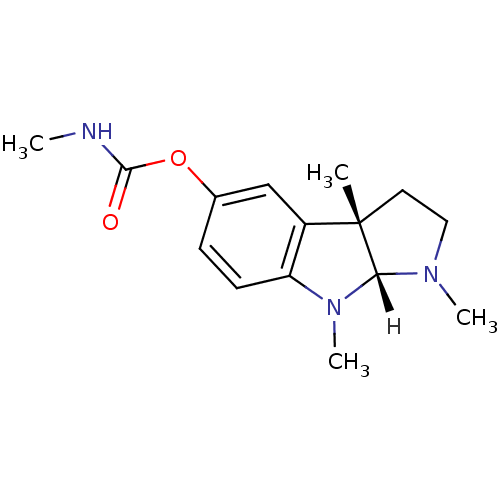
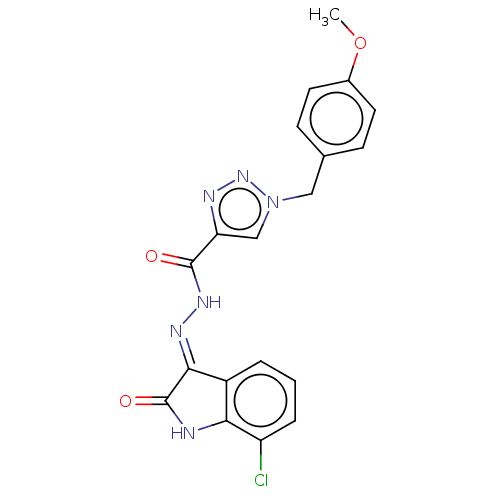
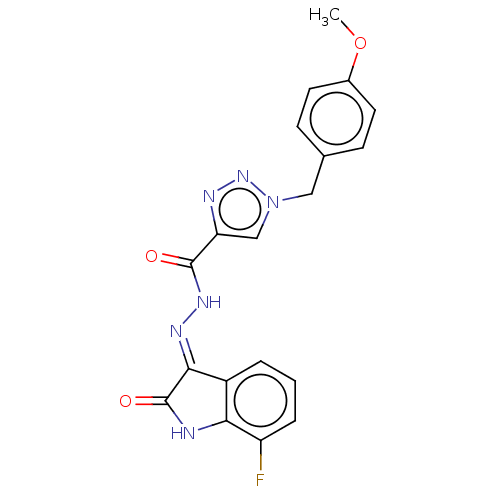
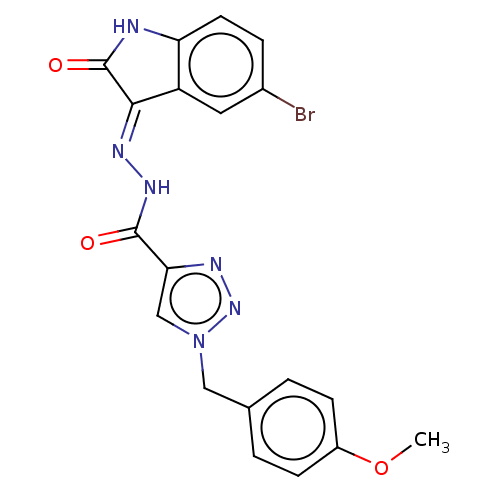
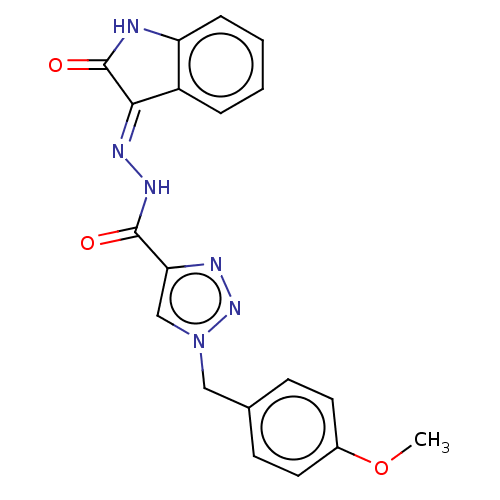
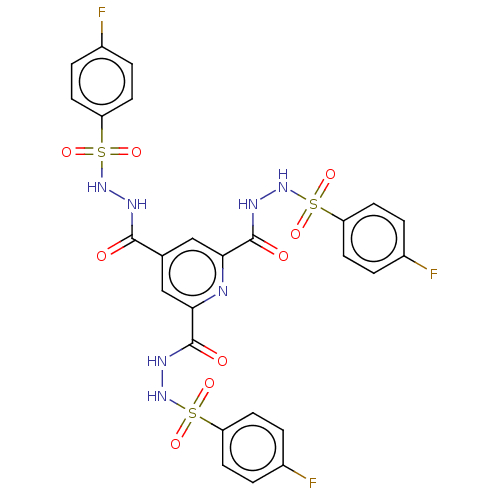
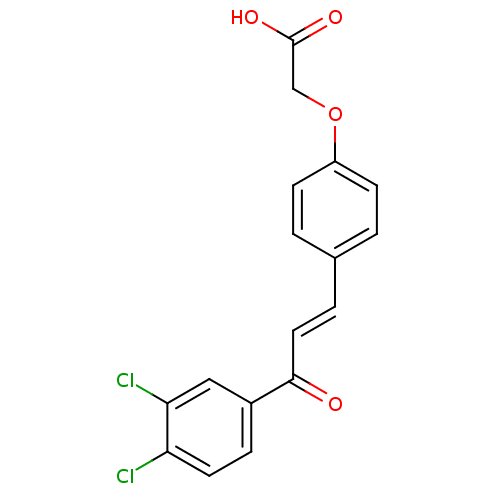
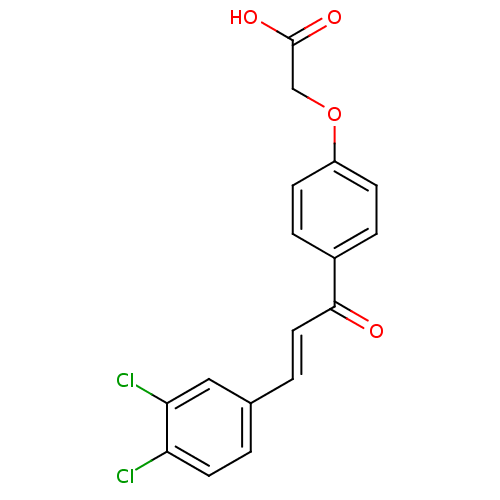
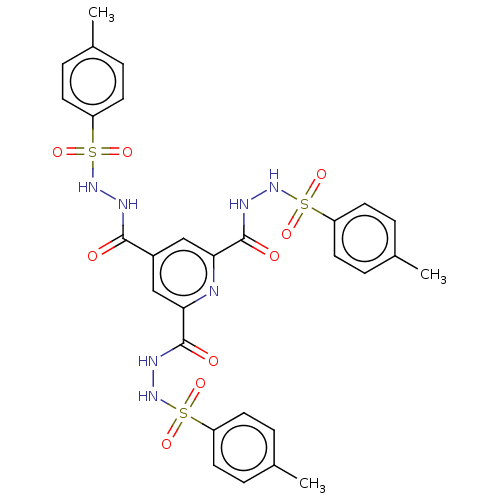
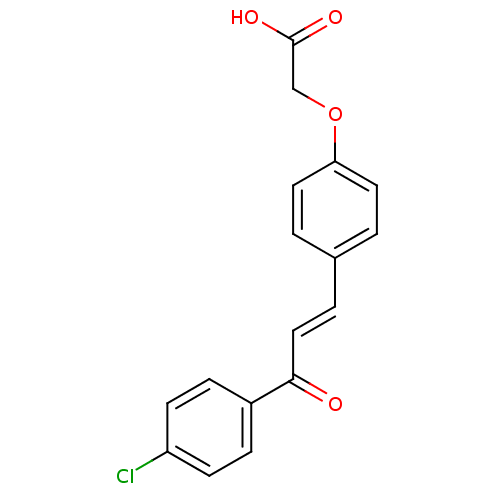
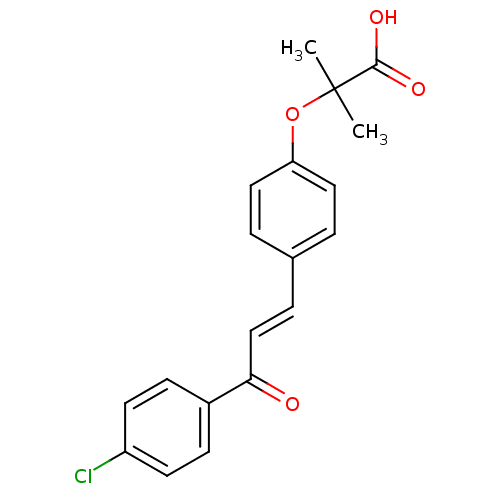
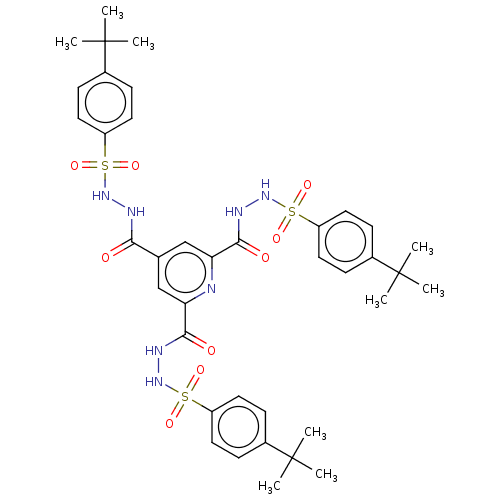
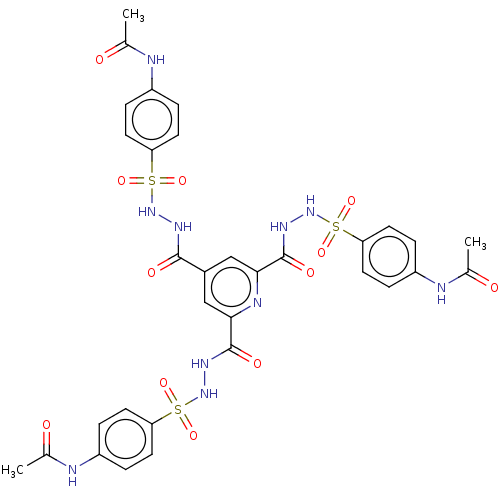
Affinity DataIC50: 40nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 850nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
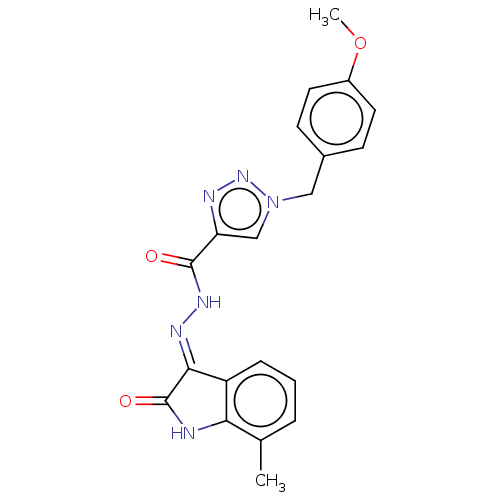
Affinity DataIC50: 1.54E+3nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
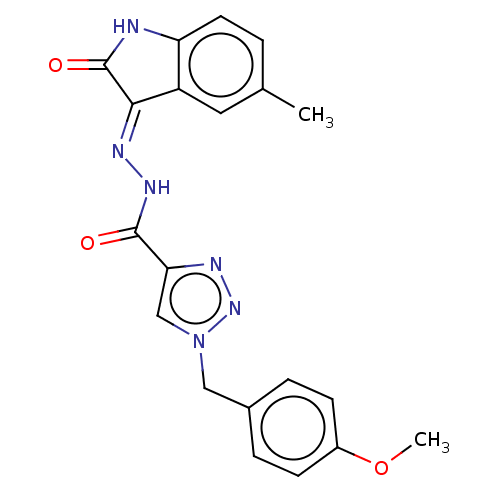
Affinity DataIC50: 1.54E+3nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
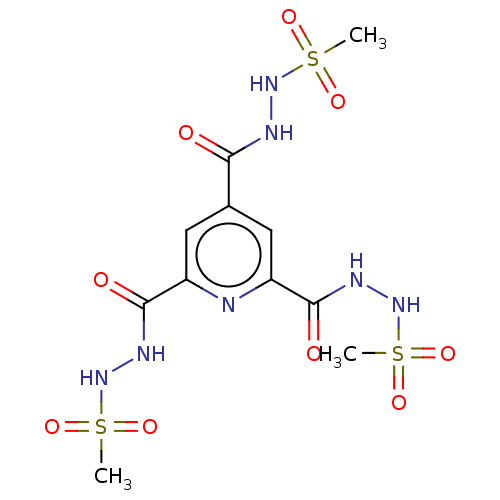
Affinity DataIC50: 8.40E+3nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 8.40E+3nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.02E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.02E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
Affinity DataIC50: 1.08E+4nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.25E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.25E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.41E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.41E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.87E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.87E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.92E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 1.92E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 2.00E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 2.00E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 2.00E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
TargetMAP/microtubule affinity-regulating kinase 4(Homo sapiens (Human))
Jamia Millia Islamia
Curated by ChEMBL
Jamia Millia Islamia
Curated by ChEMBL
Affinity DataIC50: 2.00E+4nMAssay Description:Inhibition of human recombinant MARK4 expressed in Escherichia coli M15 cells after 2 hrs in presence of [gamma-32-P]ATP by TLC analysis based ATPase...More data for this Ligand-Target Pair
Affinity DataIC50: 2.56E+4nMpH: 6.8Assay Description:The α-glucosidase inhibition activity was performed with slight modifications according to method. Total volume of 100 無 reaction mixture conta...More data for this Ligand-Target Pair
Affinity DataIC50: 2.68E+4nMpH: 6.8Assay Description:The α-glucosidase inhibition activity was performed with slight modifications according to method. Total volume of 100 無 reaction mixture conta...More data for this Ligand-Target Pair
Affinity DataIC50: 2.88E+4nMpH: 6.8Assay Description:The α-glucosidase inhibition activity was performed with slight modifications according to method. Total volume of 100 無 reaction mixture conta...More data for this Ligand-Target Pair
Affinity DataIC50: 2.91E+4nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 3.22E+4nMpH: 6.8Assay Description:The α-glucosidase inhibition activity was performed with slight modifications according to method. Total volume of 100 無 reaction mixture conta...More data for this Ligand-Target Pair
Affinity DataIC50: 3.83E+4nMpH: 6.8Assay Description:The α-glucosidase inhibition activity was performed with slight modifications according to method. Total volume of 100 無 reaction mixture conta...More data for this Ligand-Target Pair
Affinity DataIC50: 4.38E+4nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 4.48E+4nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 4.90E+4nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 5.02E+4nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 1.12E+5nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 1.17E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 1.25E+5nMpH: 6.8Assay Description:The α-glucosidase inhibition activity was performed with slight modifications according to method. Total volume of 100 無 reaction mixture conta...More data for this Ligand-Target Pair
Affinity DataIC50: 2.06E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMpH: 7.4 T: 2°CAssay Description:Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). H...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+5nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+5nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+5nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+5nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+5nMpH: 6.8Assay Description:The α-glucosidase inhibition activity was performed with slight modifications according to method. Total volume of 100 無 reaction mixture conta...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+5nMpH: 7.7Assay Description:The AChE inhibition assay was performed according to the Ellman's method. Total volume of 100 無 reaction mixture contained 60 無 50 mM Na2HPO4 b...More data for this Ligand-Target Pair
Affinity DataKd: 2.20E+5nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair
Affinity DataKd: 9.00E+4nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair
Affinity DataKd: 2.44E+5nMpH: 7.4 T: 2°CAssay Description:NMR measurements consisted of monitoring changes in chemical shifts and line widths of the backbone amide resonances of uniformly 15N-enriched MDM2 s...More data for this Ligand-Target Pair