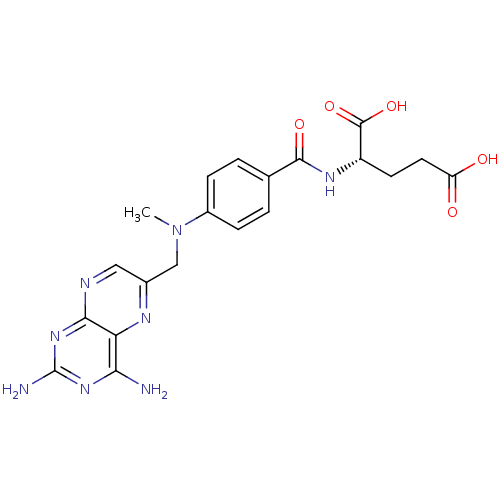
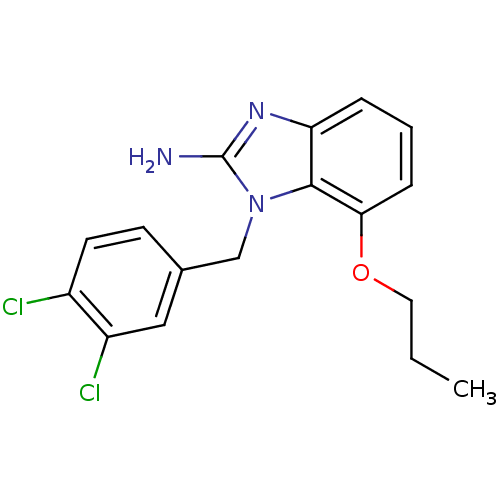
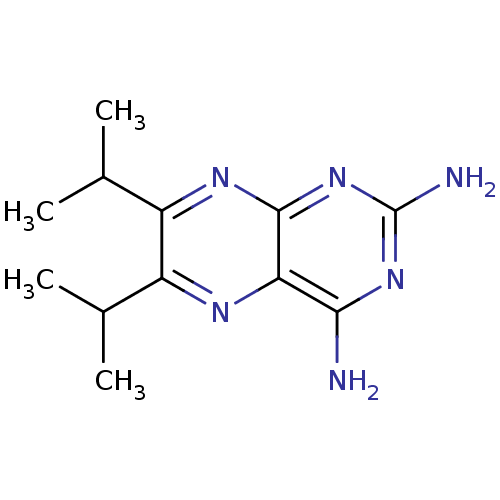
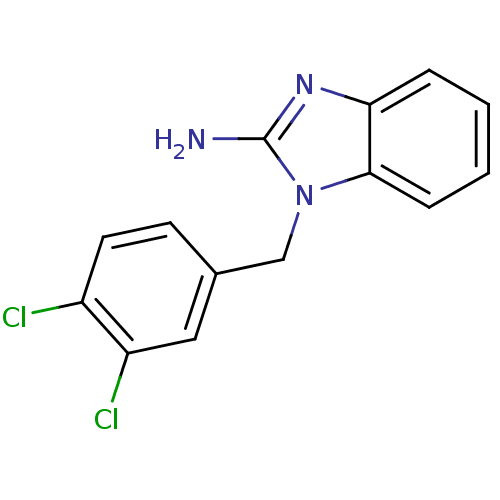
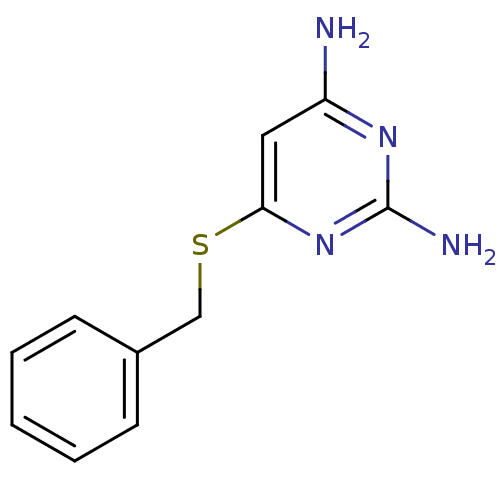
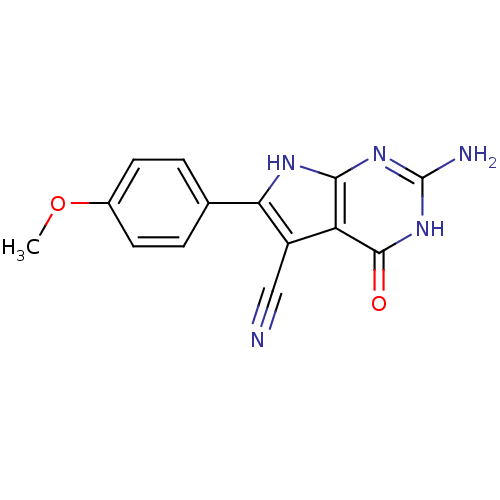
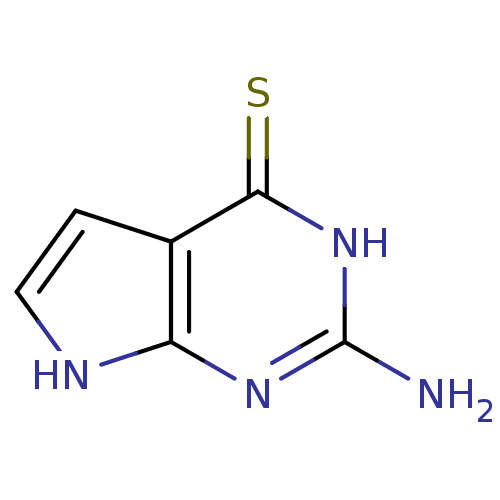
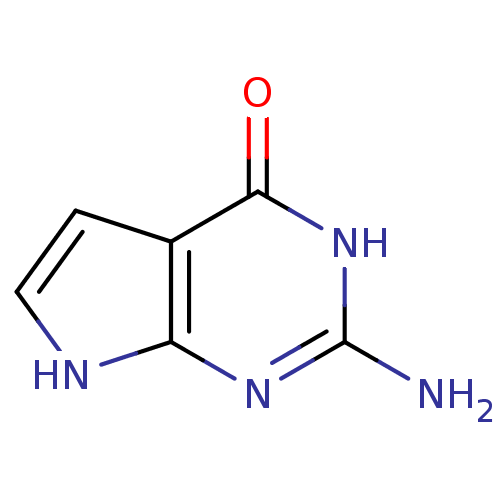
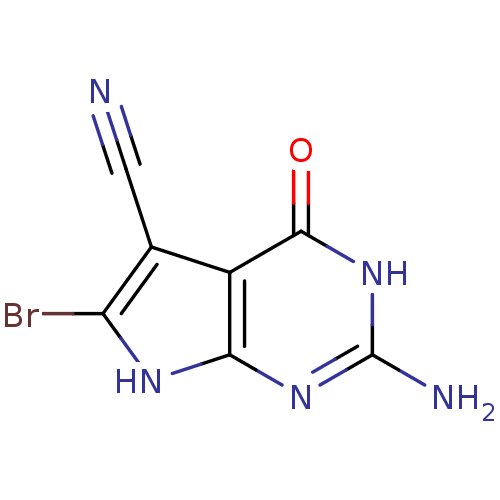
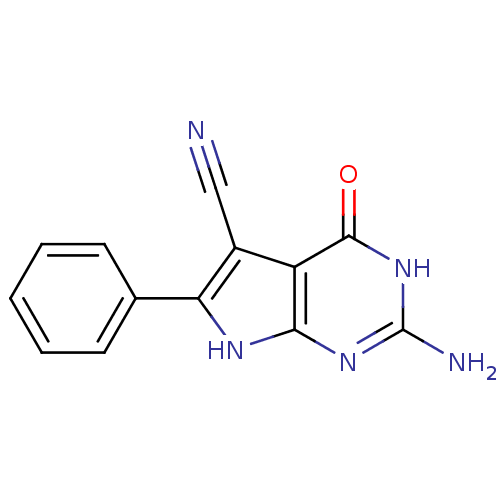
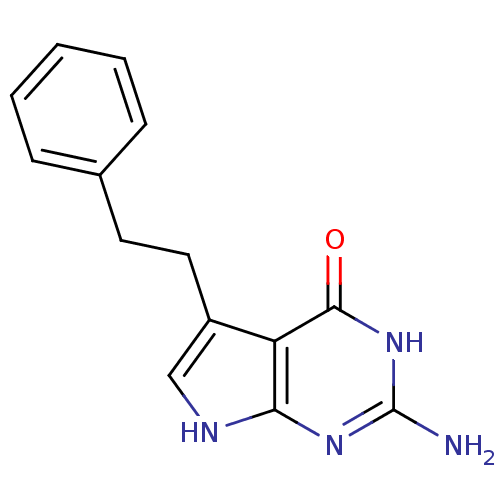
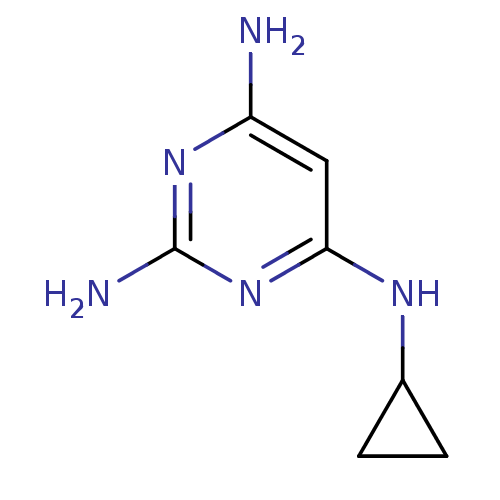
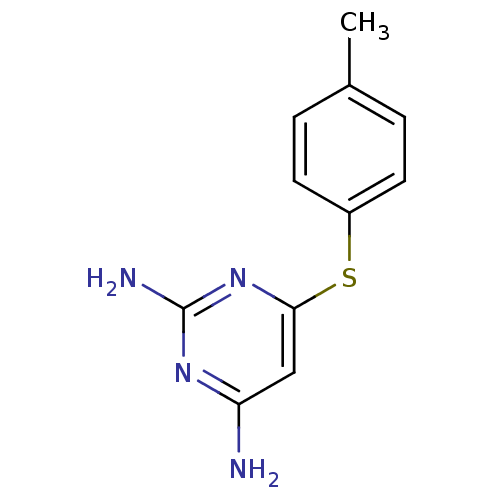
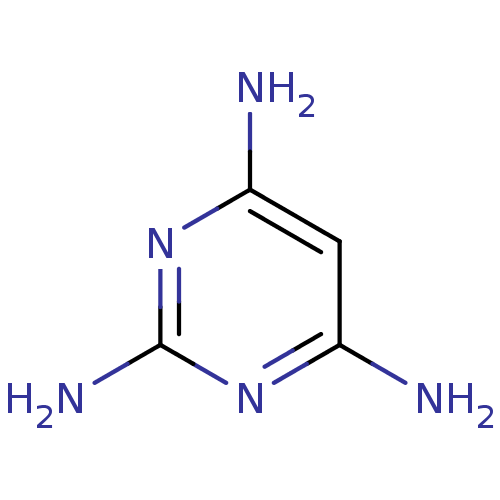
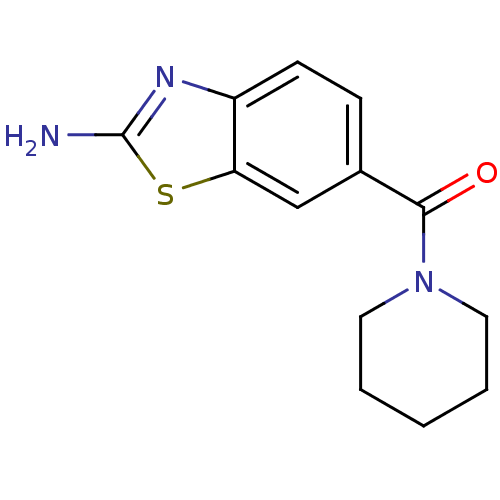
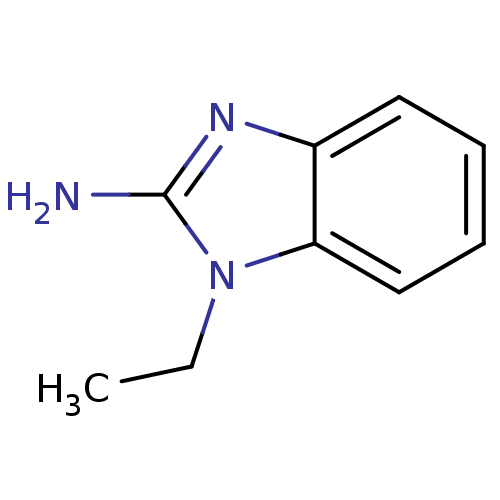
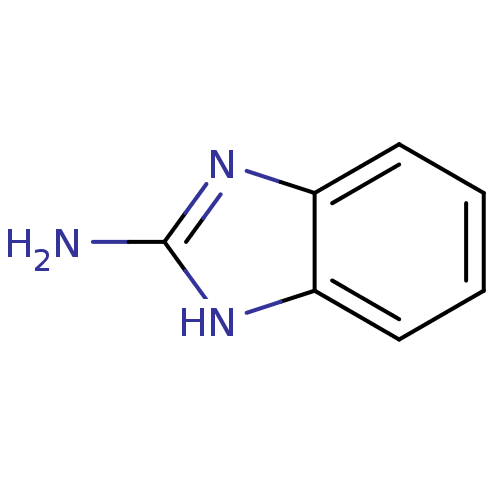
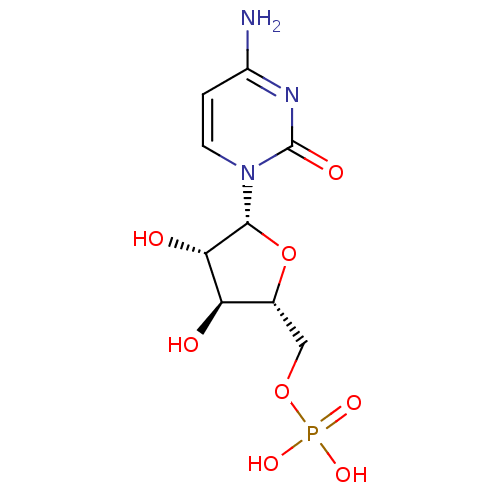
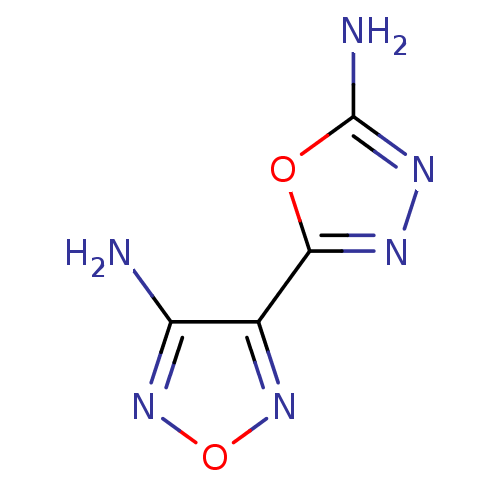
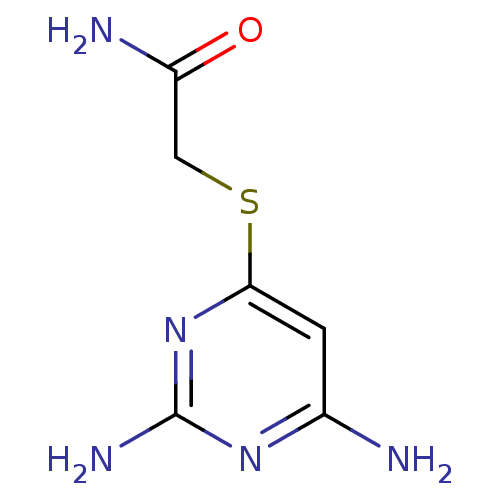
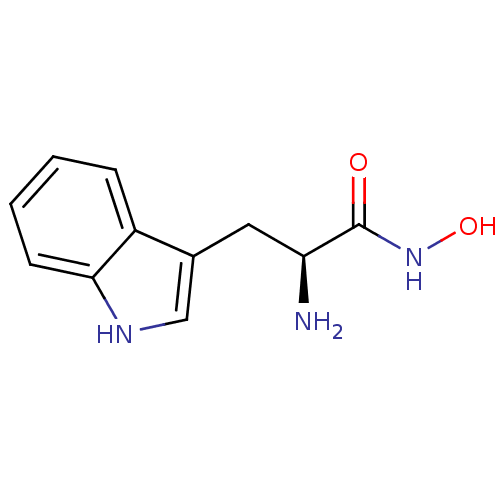
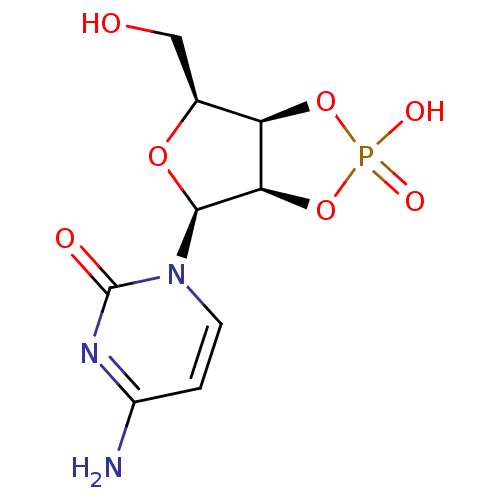
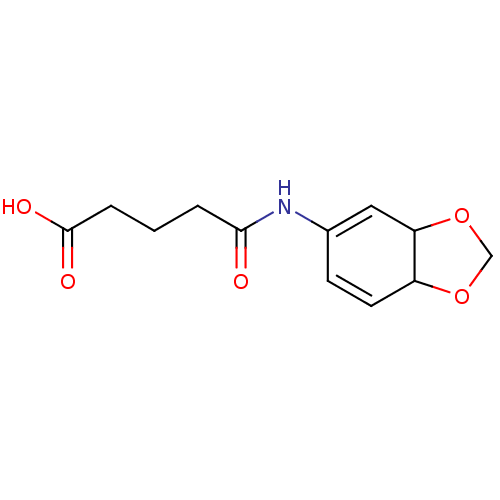
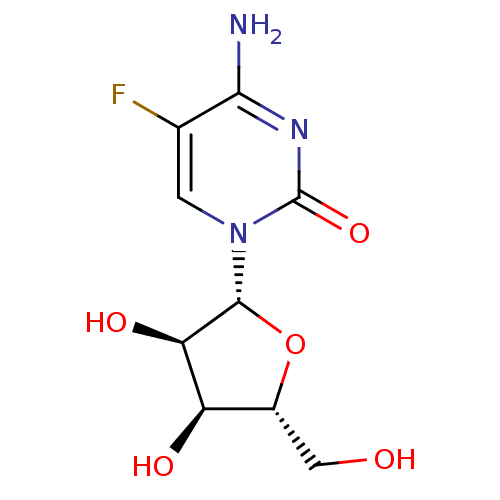
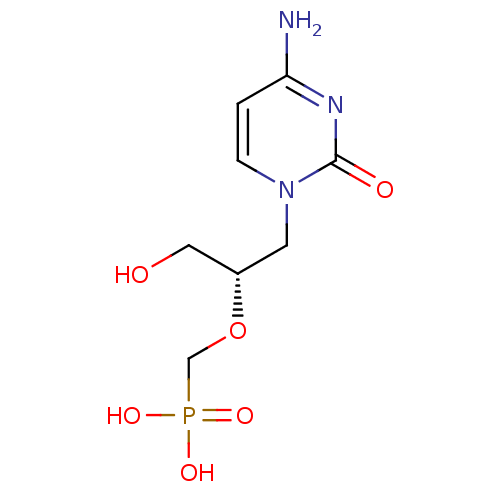
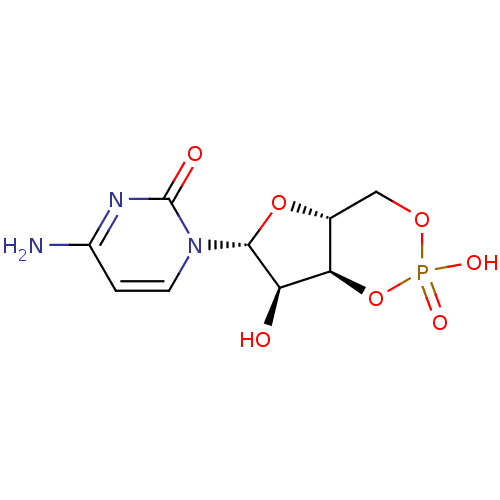
Affinity DataKi: 7nM ΔG°: -46.2kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
TargetBifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial(Homo sapiens (Human))
University of Dundee
Curated by ChEMBL
University of Dundee
Curated by ChEMBL
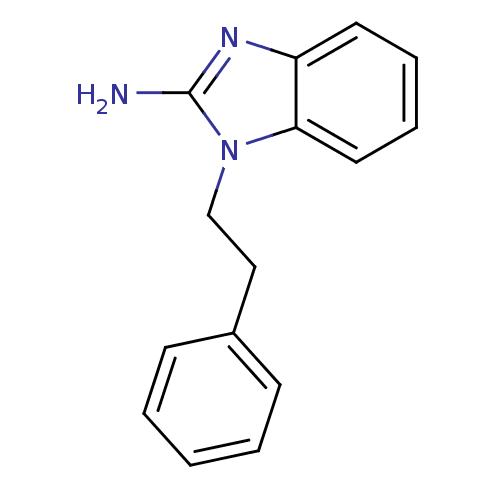
Affinity DataKi: 29nMAssay Description:Competitive inhibition of human FolD dehydrogenase activityMore data for this Ligand-Target Pair
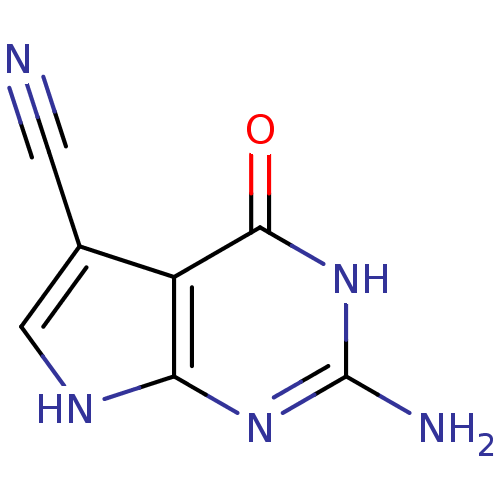
Affinity DataKi: 39nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
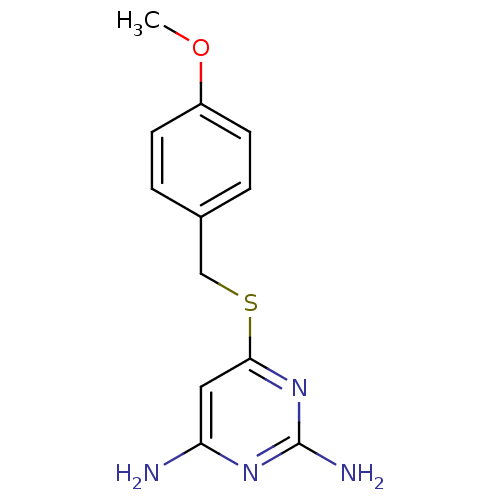
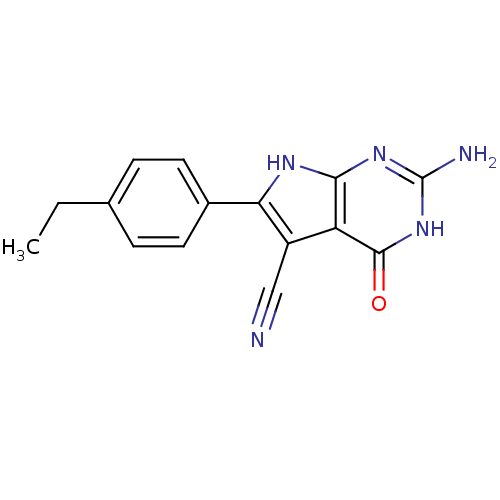
Affinity DataKi: 47nM ΔG°: -41.5kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 240nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 400nM ΔG°: -36.3kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 510nM ΔG°: -35.7kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 600nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 2.60E+3nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 2.70E+3nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 3.40E+3nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 3.40E+3nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 4.20E+3nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 9.80E+3nM ΔG°: -28.4kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 1.06E+4nM ΔG°: -28.2kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 1.20E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 1.64E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 2.11E+4nM ΔG°: -26.5kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 2.39E+4nM ΔG°: -26.2kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: >2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: >2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: >2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: >2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: >2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: >2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: >2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: >2.70E+4nMAssay Description:Inhibition of Leishmania major recombinant PTR1More data for this Ligand-Target Pair
Affinity DataKi: 1.41E+5nM ΔG°: -21.8kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: >2.00E+5nM ΔG°: >-21.0kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Affinity DataKi: 2.88E+5nM ΔG°: -20.1kJ/molepH: 6.0 T: 2°CAssay Description:TbPTR1 activity was measured in 96-well microtiter plates via reduction of cytochrome c (cytc) as a result of the enzymatic production of tetrahydrob...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DataKd: 1.89E+7nMpH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DatapH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DatapH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DataKd: 1.92E+3nMpH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DataKd: 2.05E+7nMpH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DatapH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DataKd: 2.02E+6nMpH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DataKd: 4.00E+7nMpH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair
Target2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase(Escherichia coli (strain K12))
University of Dundee
University of Dundee
Affinity DataKd: 9.00E+5nMpH: 7.0 T: 2°CAssay Description:An SPR assay was used to determine the binding affinities of compounds. Experiments were performed on a Biacore 3000 (Biacore, Uppsala, Sweden) instr...More data for this Ligand-Target Pair


 3D Structure (crystal)
3D Structure (crystal)