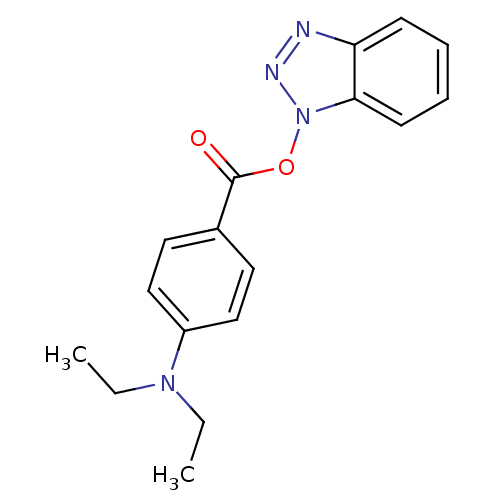
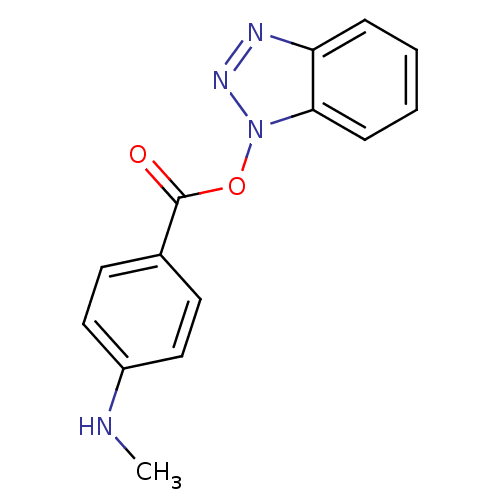
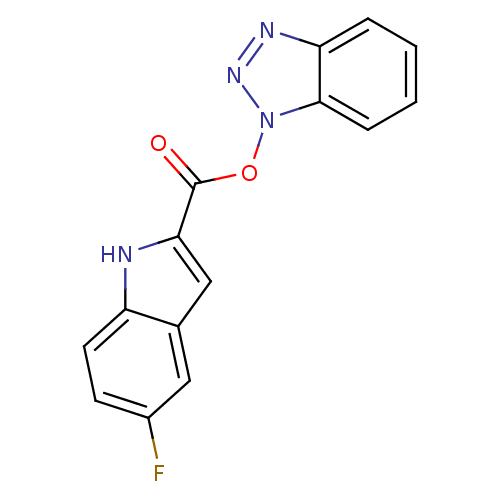
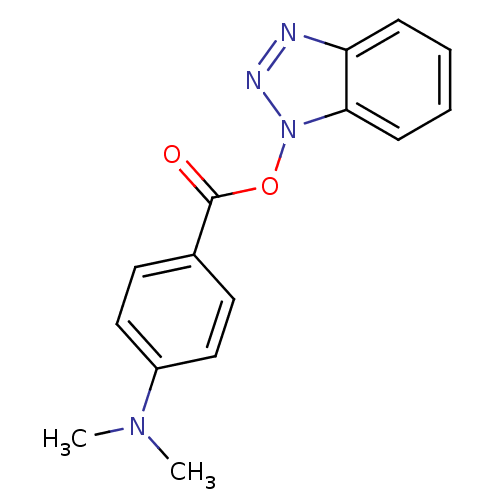
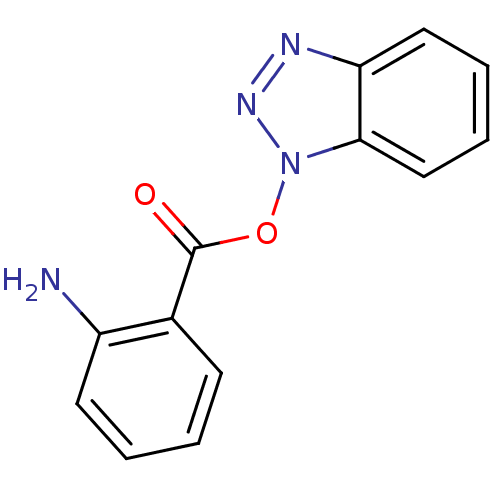
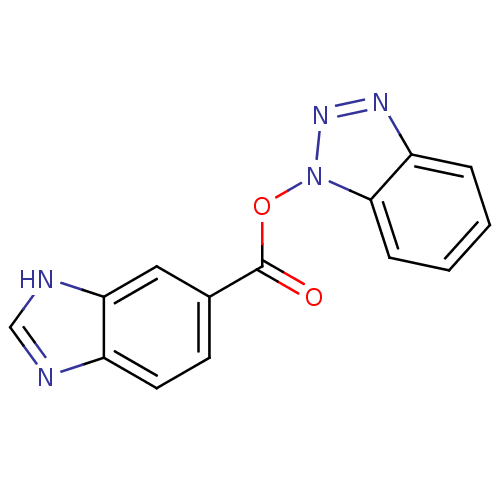
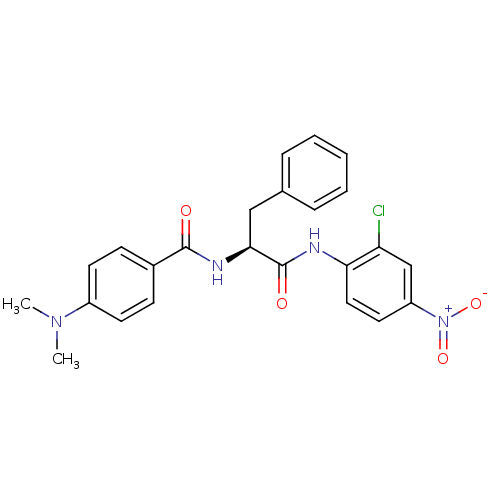
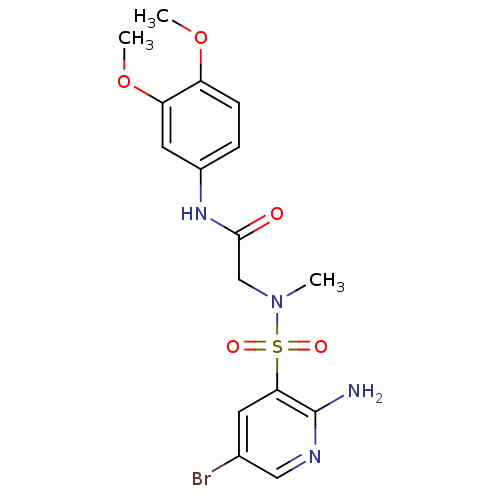
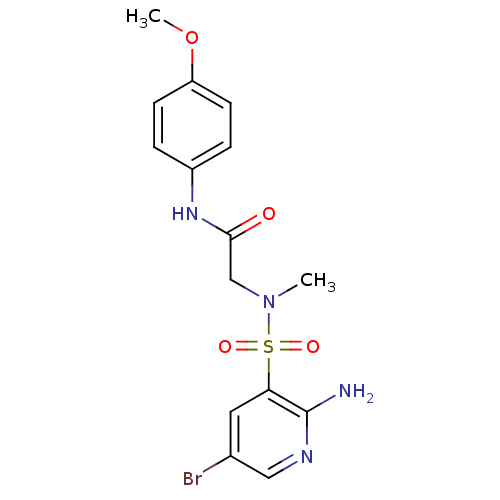
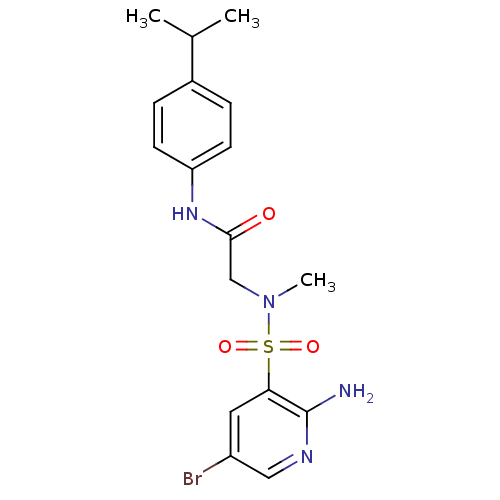
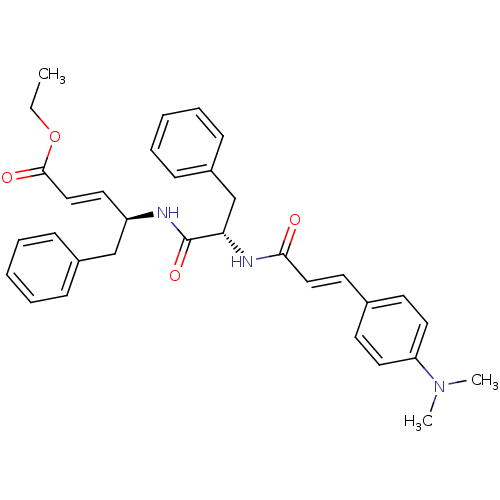
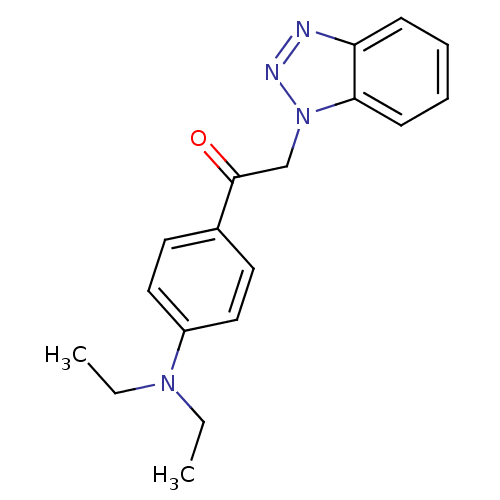
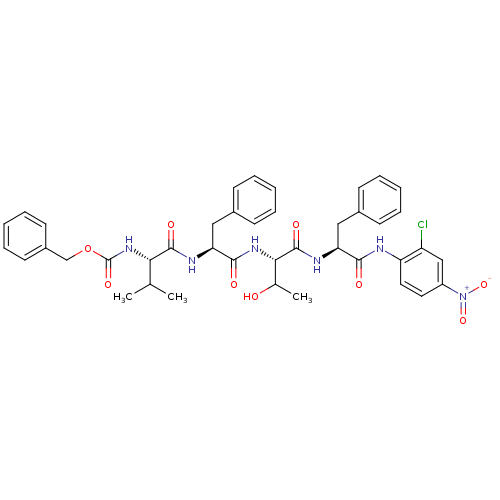
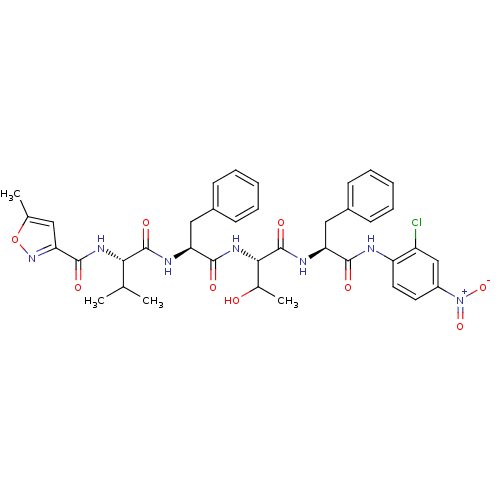
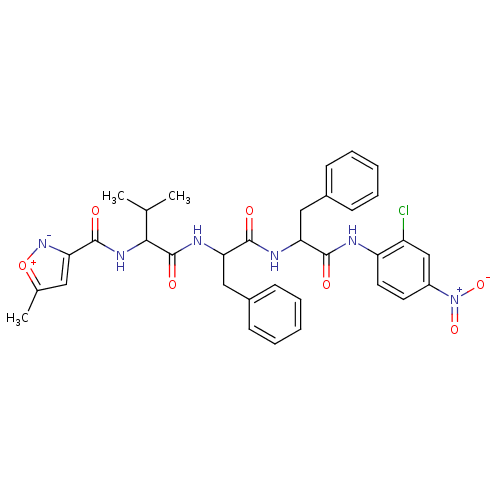
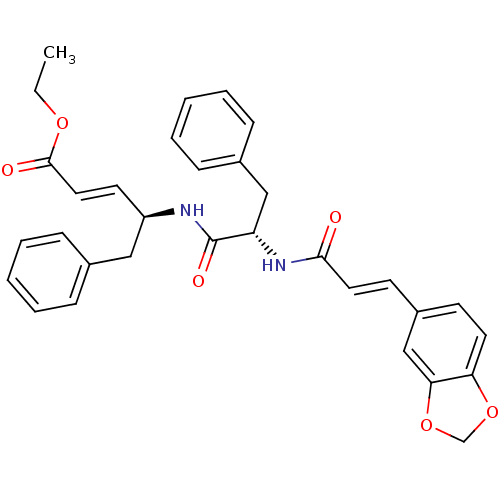
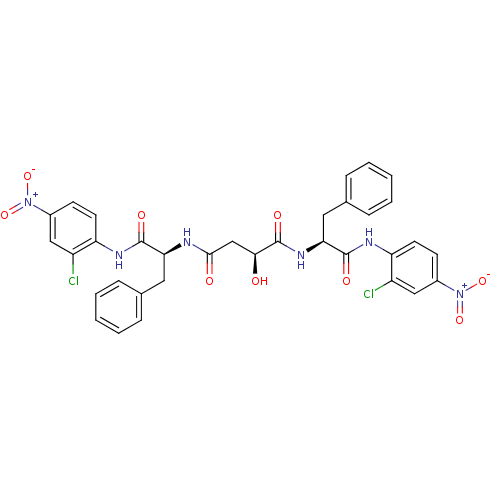
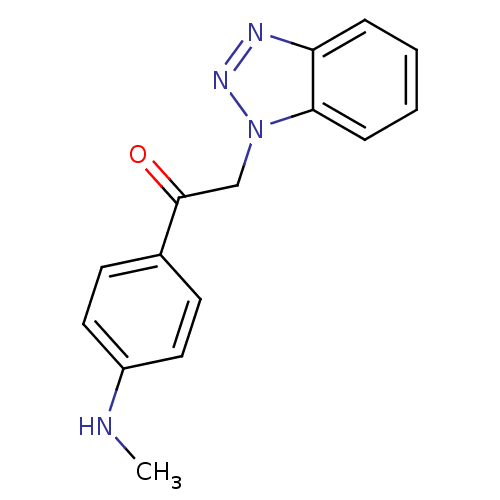
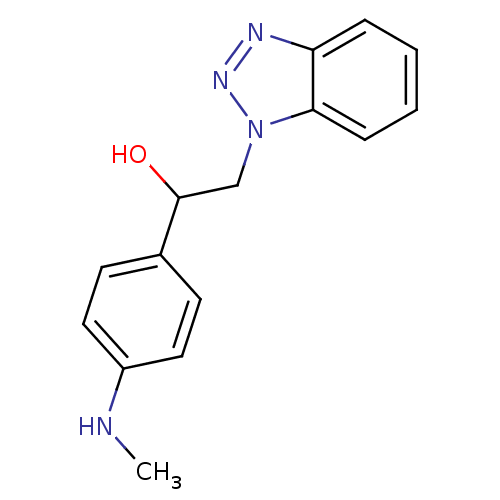
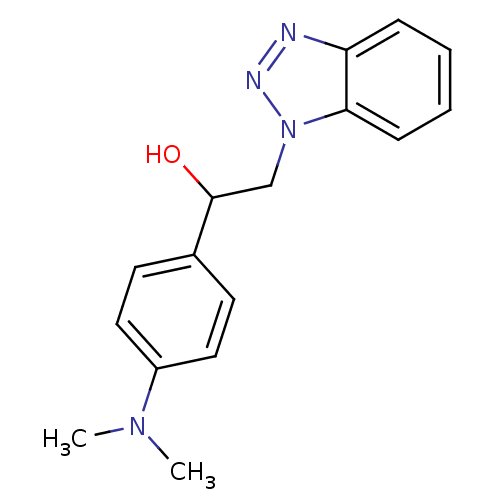
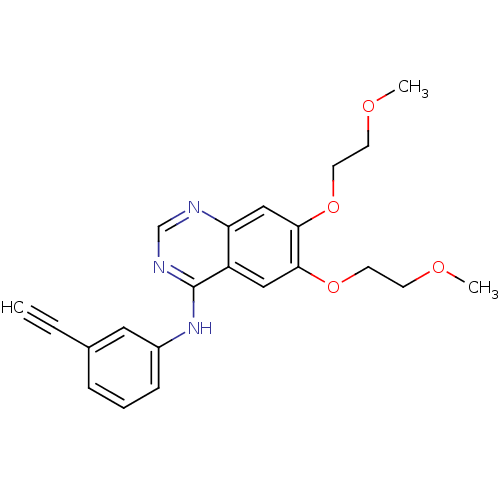
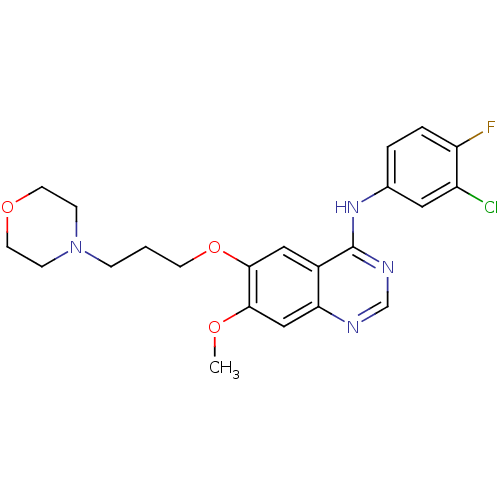
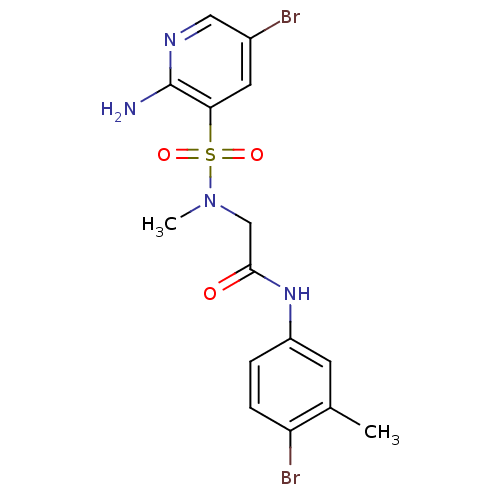
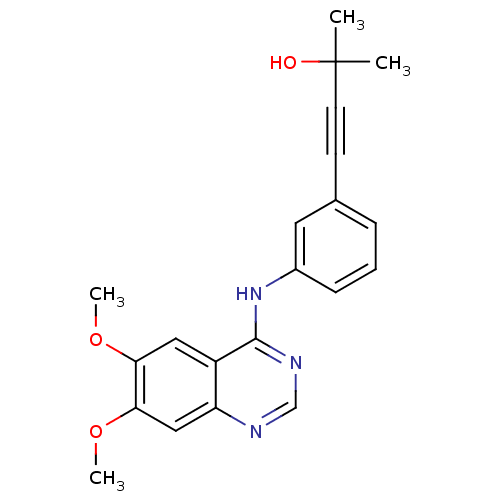
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 7.5nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 11.1nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 12.1nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 12.3nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 13.8nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 17.4nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 19.5nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 22.9nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 30nM ΔG°: -42.9kJ/mole IC50: 60nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
Affinity DataKi: 37nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 78nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 93nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 140nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 143nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 155nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 155nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 233nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 289nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
Affinity DataKi: 333nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 520nM ΔG°: -35.9kJ/mole IC50: 1.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 1.00E+3nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 1.51E+3nM ΔG°: -33.2kJ/mole IC50: 6.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 1.61E+3nM ΔG°: -33.1kJ/mole IC50: 5.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 2.29E+3nM ΔG°: -32.2kJ/mole IC50: 5.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 2.48E+3nM ΔG°: -32.0kJ/mole IC50: 5.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 2.90E+3nM ΔG°: -31.6kJ/mole IC50: 7.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 3.05E+3nM ΔG°: -31.5kJ/mole IC50: 7.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 3.10E+3nM ΔG°: -31.4kJ/mole IC50: 4.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 4.30E+3nM ΔG°: -30.6kJ/mole IC50: 5.00E+3nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 4.50E+3nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 6.44E+3nM ΔG°: -29.6kJ/mole IC50: 1.00E+4nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 6.70E+3nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: 9.05E+3nM ΔG°: -28.8kJ/mole IC50: 1.00E+4nMpH: 7.0 T: 2°CAssay Description:The effects of compound on enzyme activity were measured by using a fluorogenic peptide cleavage assay. Enhanced fluorescence caused by cleavage of t...More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: >5.00E+4nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Academia Sinica
Academia Sinica
Affinity DataKi: >5.00E+4nMpH: 7.5Assay Description:Inhibition assay against SARS-CoV 3CL protease, a fluorometric assay was utilized to determine the inhibition constants of the samples.More data for this Ligand-Target Pair
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
In DepthDetails
Article
BindingDB Entry DOI: 10.7270/Q25Q4VT1PubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)
BindingDB Entry DOI: 10.7270/Q25Q4VT1PubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
In DepthDetails
Article
BindingDB Entry DOI: 10.7270/Q25Q4VT1PubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)
BindingDB Entry DOI: 10.7270/Q25Q4VT1PubMedDrugBank
MMDB
PDB
 3D Structure (crystal)
3D Structure (crystal)TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
Affinity DataIC50: 100nMAssay Description:IC50 values were determined with serial dilutions (from 100 to 0.1 uM) of inhibitors by a spectrophotometric assay with DTNB, lipoamide, and NADH or ...More data for this Ligand-Target Pair
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL
TargetEpidermal growth factor receptor(Homo sapiens (Human))
Development Center for Biotechnology
Curated by ChEMBL
Development Center for Biotechnology
Curated by ChEMBL