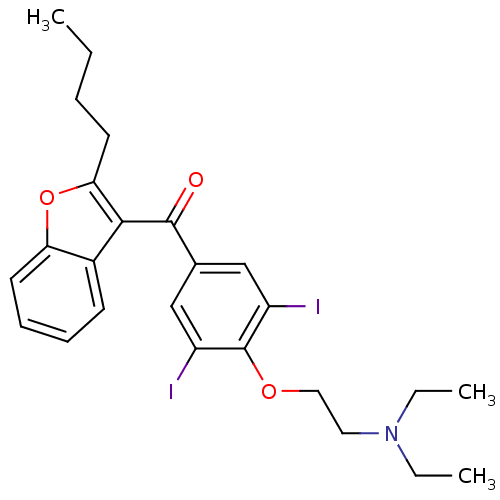
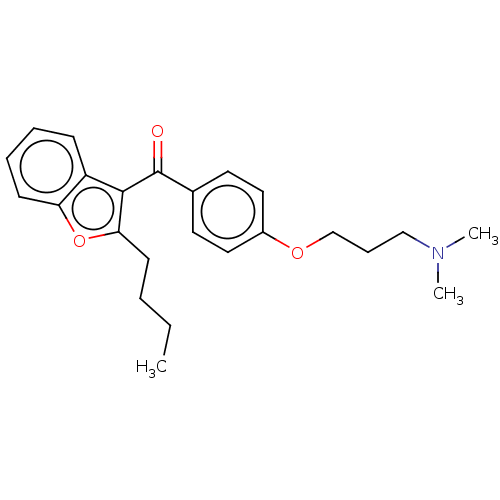
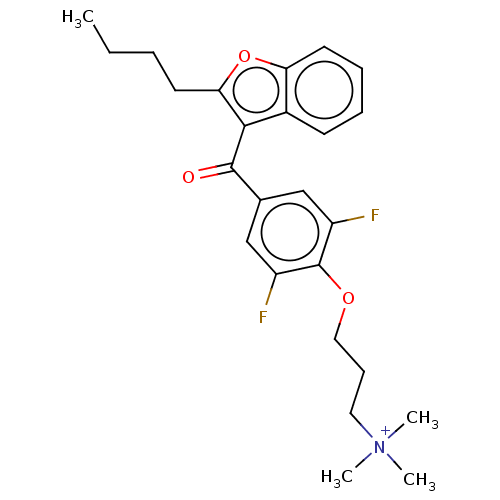
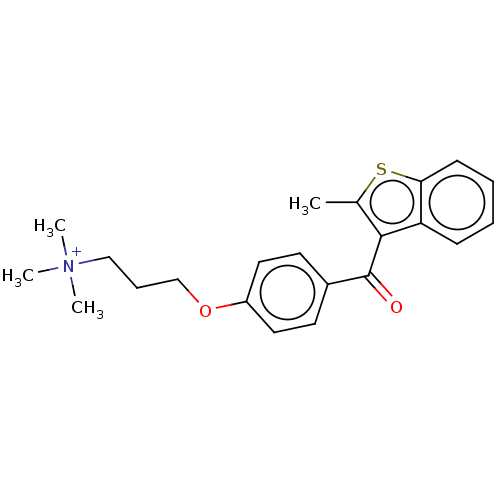
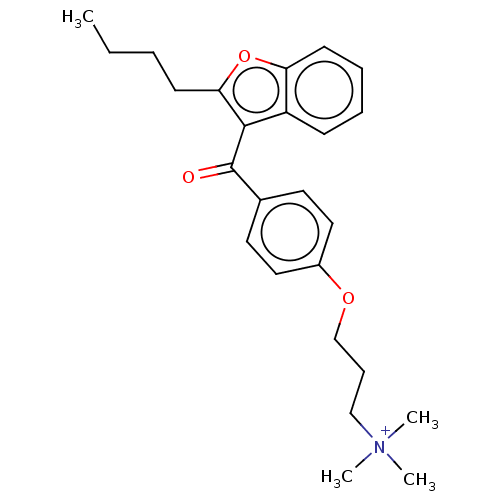
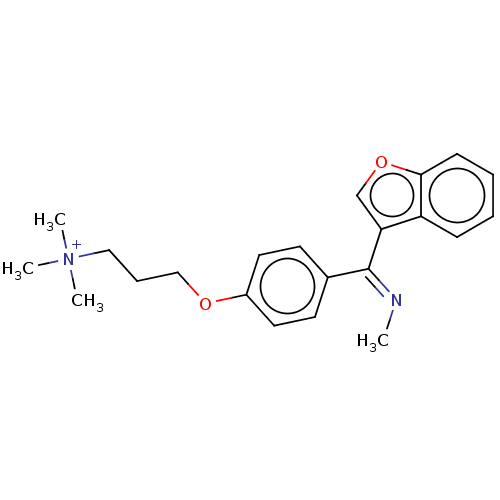
Affinity DataIC50: 2.70E+3nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 8.40E+4nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 1.58E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 2.20E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 2.28E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 3.34E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 3.59E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 5.01E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 5.33E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 5.79E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 5.98E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 6.82E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 7.40E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 9.76E+5nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair
Affinity DataIC50: 1.29E+6nMAssay Description:Displacement of C-terminally biotinylated-H3K4me3 (1 to 21 residues) peptide from KDM7C (PHD) (unknown origin) preincubated for 15 mins followed by p...More data for this Ligand-Target Pair