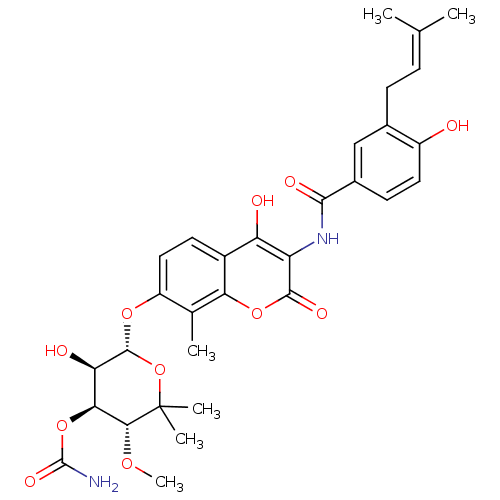
Affinity DataKd: 1.10E+5nMpH: 6.0Assay Description:Binding of inhibitors and substrates to immobilized AcrB.More data for this Ligand-Target Pair
Affinity DataKd: 1.03E+4nMpH: 8.4Assay Description:Binding affinity to Escherichia coli K-12 multidrug efflux protein AcrB at pH 8.4 by equilibrium dialysis methodMore data for this Ligand-Target Pair
Affinity DataKd: 5.30E+5nMpH: 7.5Assay Description:Binding of inhibitors and substrates to immobilized AcrB.More data for this Ligand-Target Pair
Affinity DataKd: 5.90E+5nMpH: 7.5Assay Description:Binding of inhibitors and substrates to immobilized AcrB.More data for this Ligand-Target Pair