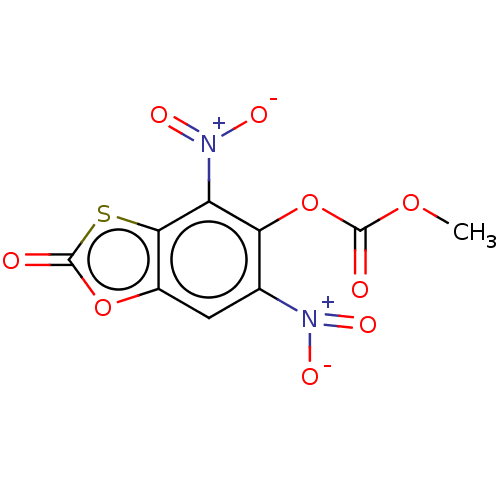
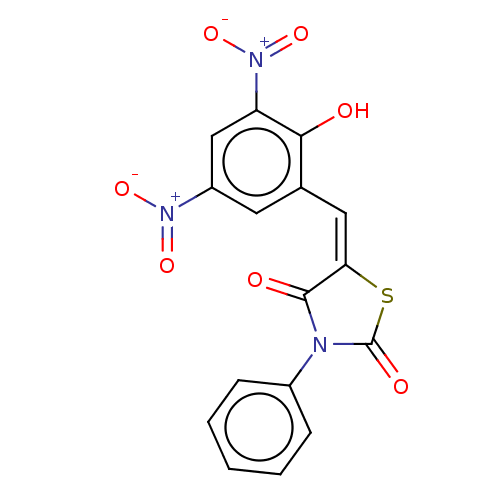
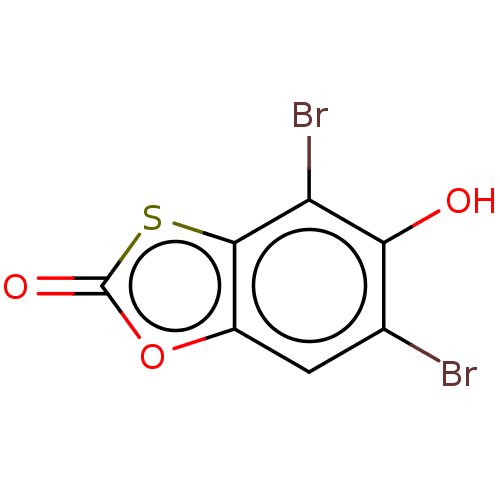
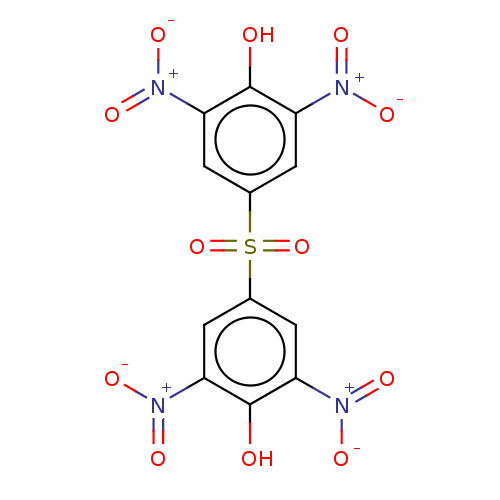
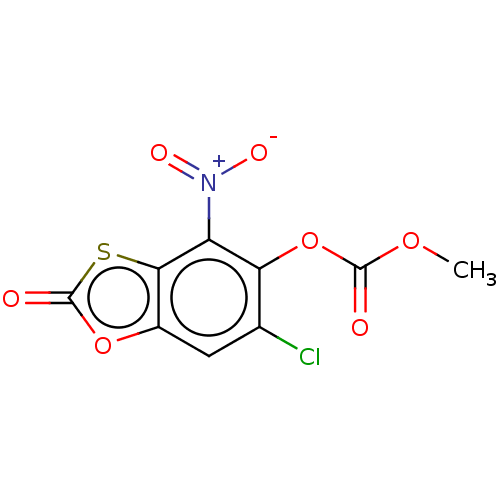
Affinity DataKi: 800nMAssay Description:Competitive inhibition of Yersinia enterocolitica Irp9 using chorismate as substrateMore data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+3nMAssay Description:Inhibition of Yersinia enterocolitica Irp9 using chorismate as substrate assessed as pyruvate production after 10 mins by microplate reader analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of Yersinia enterocolitica Irp9 using chorismate as substrate assessed as salicylate production by fluorescence spectrometryMore data for this Ligand-Target Pair
Affinity DataIC50: 4.20E+4nMAssay Description:Inhibition of Yersinia enterocolitica Irp9 using chorismate as substrate assessed as salicylate production by fluorescence spectrometryMore data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+4nMAssay Description:Inhibition of Yersinia enterocolitica Irp9 using chorismate as substrate assessed as salicylate production by fluorescence spectrometryMore data for this Ligand-Target Pair
Affinity DataIC50: 8.00E+4nMAssay Description:Inhibition of Yersinia enterocolitica Irp9 using chorismate as substrate assessed as salicylate production by fluorescence spectrometryMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+5nMAssay Description:Inhibition of Yersinia enterocolitica Irp9 using chorismate as substrate assessed as pyruvate production after 10 mins by microplate reader analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+5nMAssay Description:Inhibition of Yersinia enterocolitica Irp9 using chorismate as substrate assessed as salicylate production by fluorescence spectrometryMore data for this Ligand-Target Pair