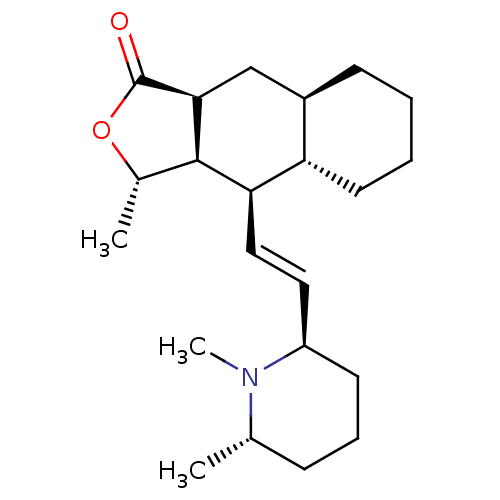
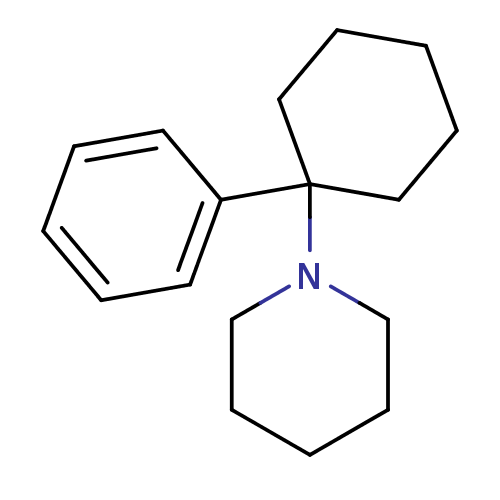
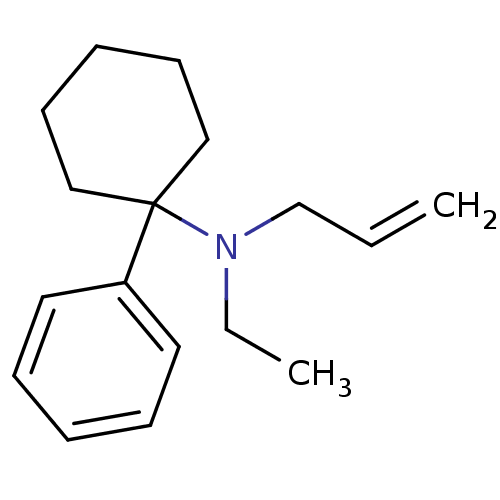
Affinity DataKd: 11nMAssay Description:Potency of the compound assessed to inhibit cAMP levels in N1E-115 neuroblastoma cells, for blocking oxotremorine-M in functional assay for Muscarini...More data for this Ligand-Target Pair
Affinity DataKd: 1.50E+4nMAssay Description:In vitro inhibition of Angiotensin I converting enzymeMore data for this Ligand-Target Pair
Affinity DataKd: 9.10E+3nMAssay Description:Displacement of [3H]5-HT binding to serotonin receptor of rat fundus membranesMore data for this Ligand-Target Pair
Affinity DataKd: 1.40E+4nMAssay Description:Displacement of [3H]5-HT binding to serotonin receptor of rat fundus membranesMore data for this Ligand-Target Pair
Affinity DataKd: 7.00E+4nMAssay Description:Displacement of [3H]5-HT binding to serotonin receptor of rat fundus membranesMore data for this Ligand-Target Pair
Affinity DataKd: 2.50E+4nMAssay Description:Displacement of [3H]5-HT binding to serotonin receptor of rat fundus membranesMore data for this Ligand-Target Pair