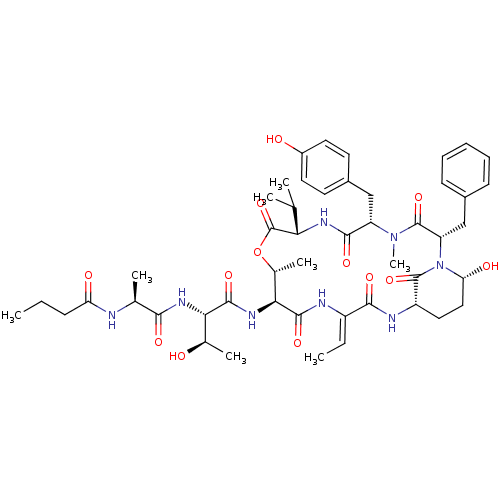
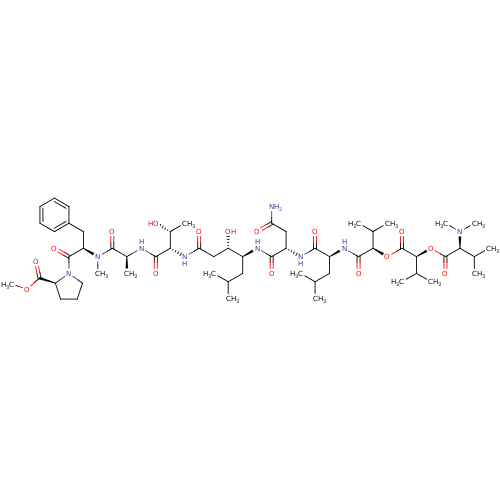
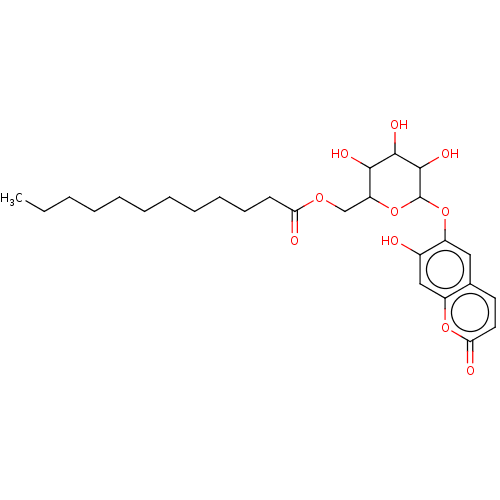
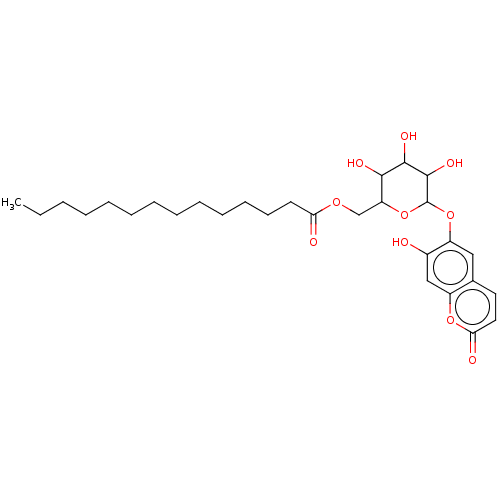
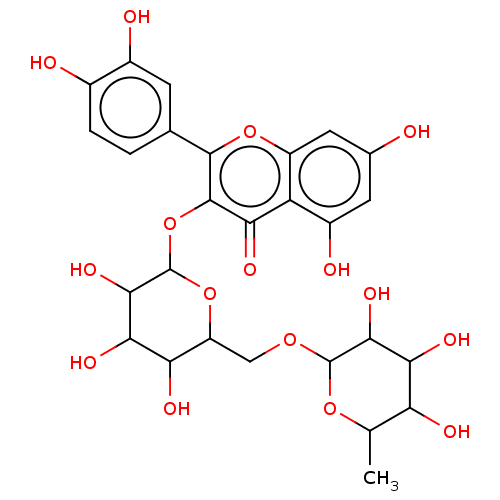
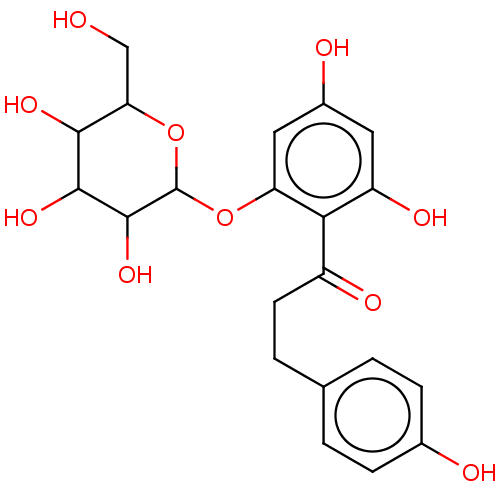
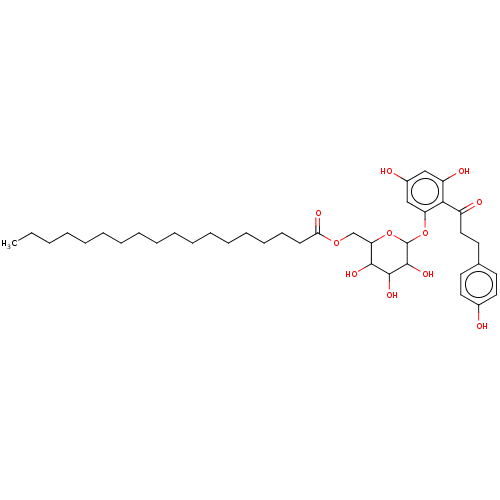
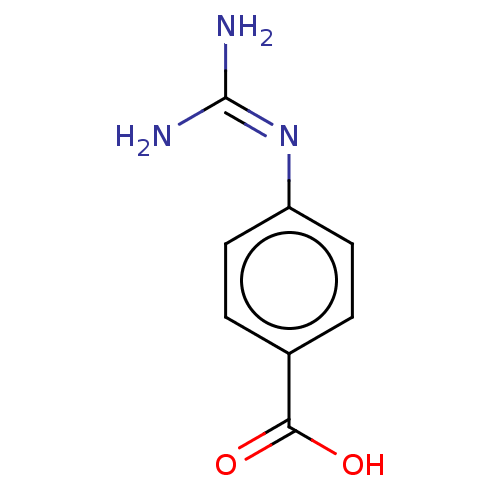
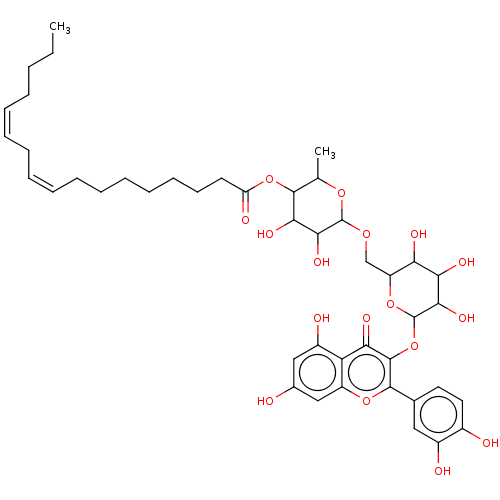
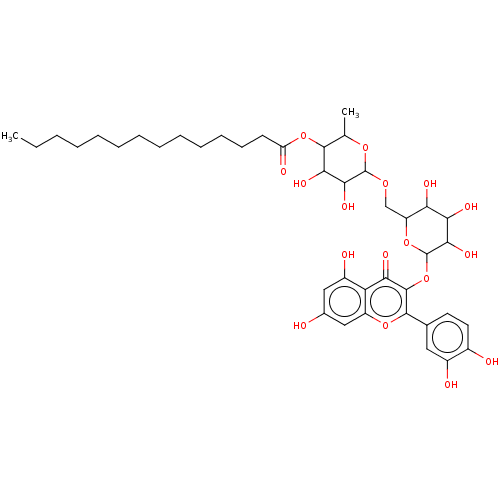
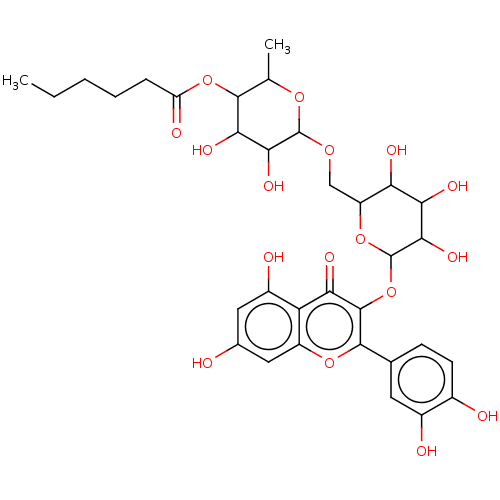
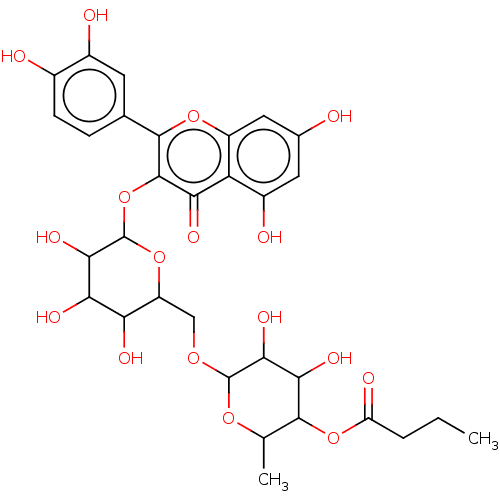
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 32nMAssay Description:Inhibition of pig pancreatic elastase preincubated for 15 mins by spectrophotometryMore data for this Ligand-Target Pair
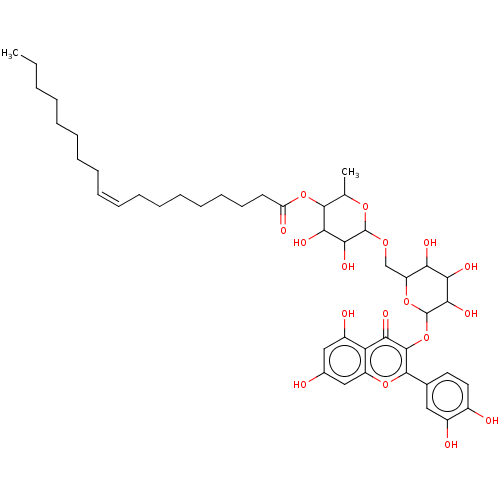
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 1.00E+4nMAssay Description:Inhibition of porcine pancreatic elastase after 10 to 15 mins by fluorescence assayMore data for this Ligand-Target Pair
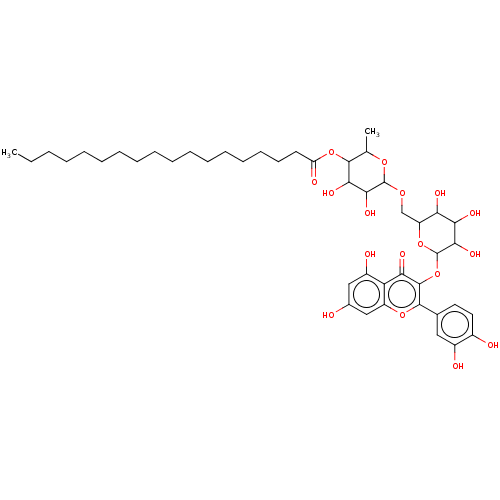
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 2.00E+4nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
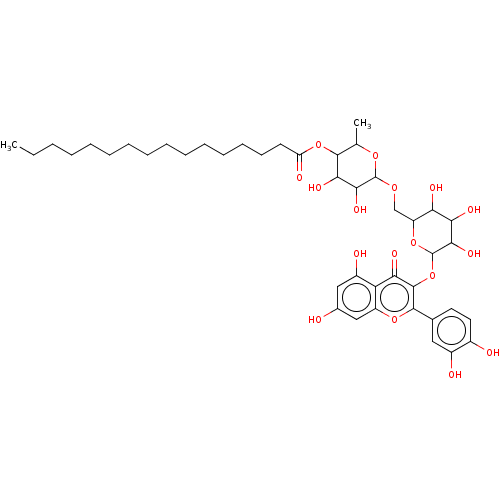
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 2.50E+4nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+4nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 6.00E+4nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 1.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 1.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 1.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair
TargetChymotrypsin-like elastase family member 1(Sus scrofa (Pig))
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Smithsonian Marine Station at Ft. Pierce
Curated by ChEMBL
Affinity DataIC50: 5.00E+5nMpH: 8.0Assay Description:The rate of hydrolysis of the substrates (0.3 mM) by trypsin (18 U/ml), thrombin (12.5 NIH U/ml), urokinase (62500 IU/ml) and elastase (8.4 U/ml) was...More data for this Ligand-Target Pair