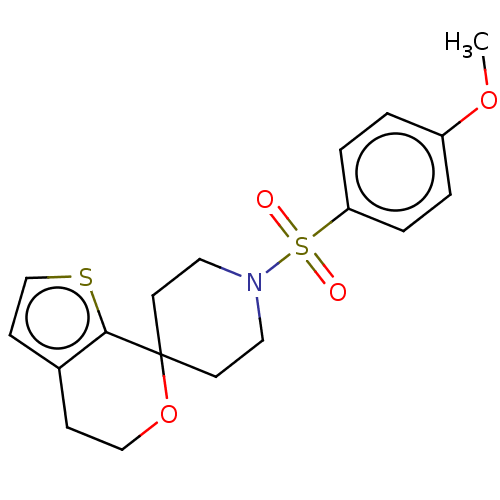
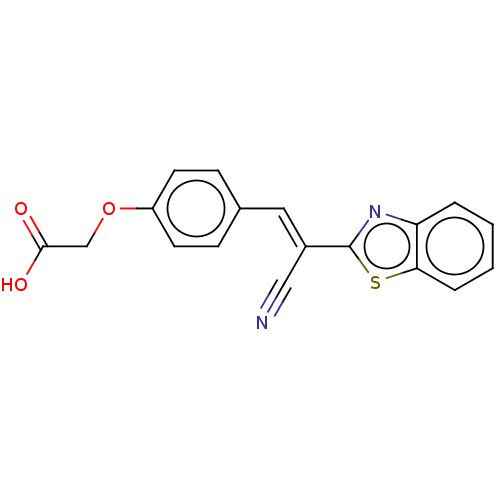
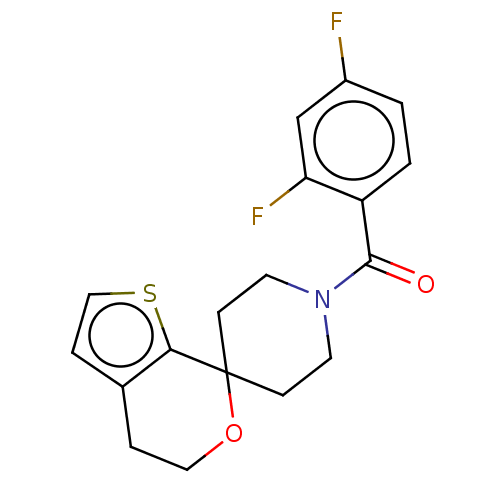
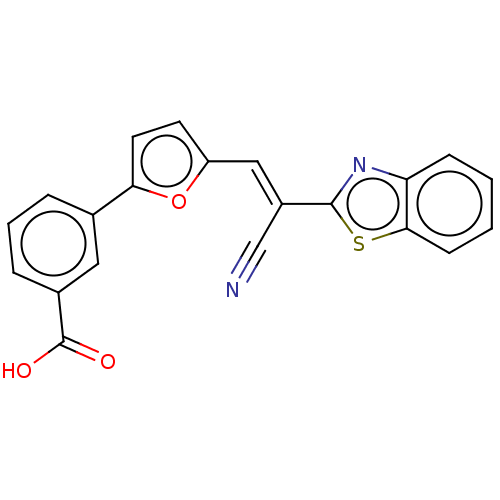
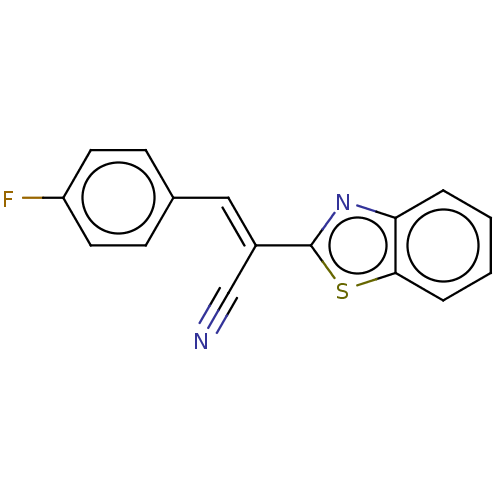
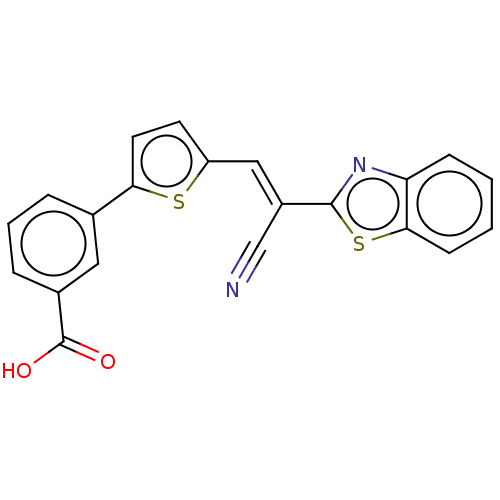
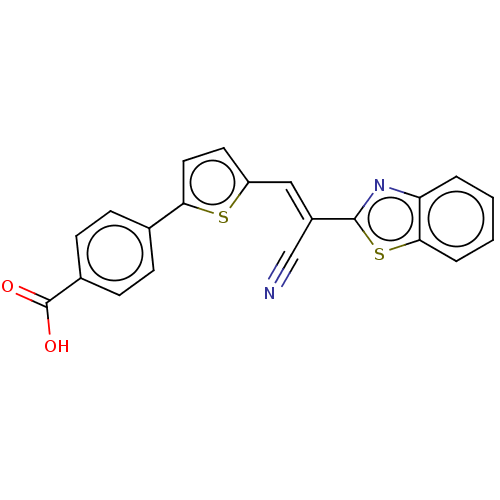
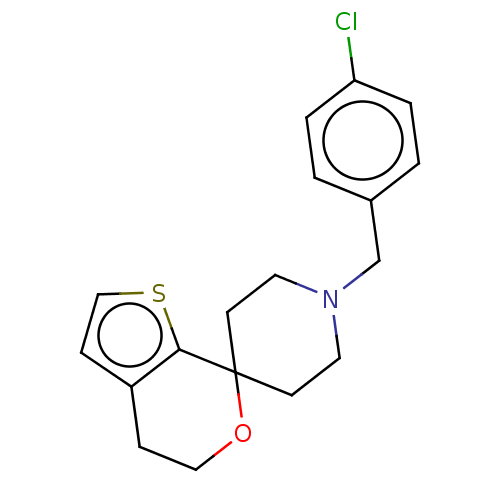
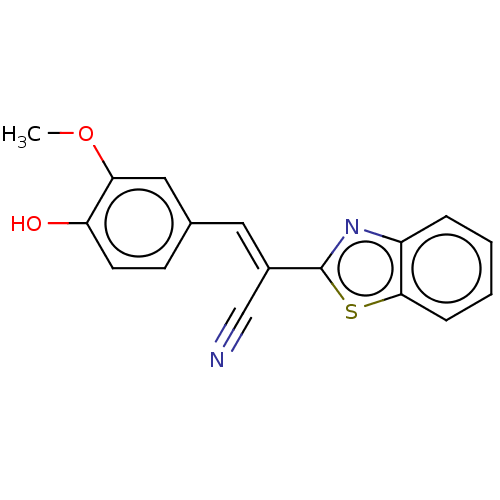
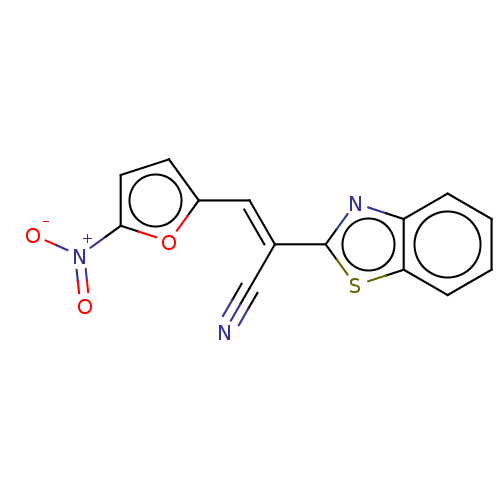
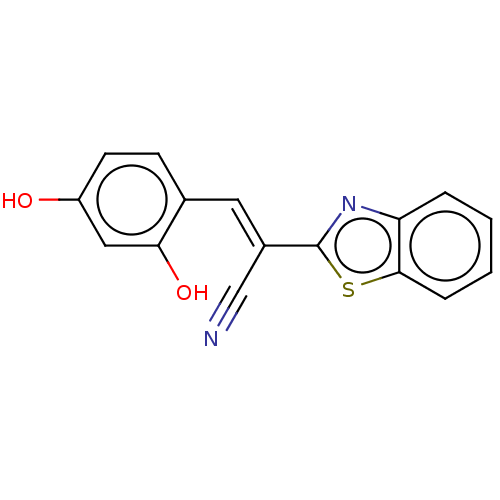
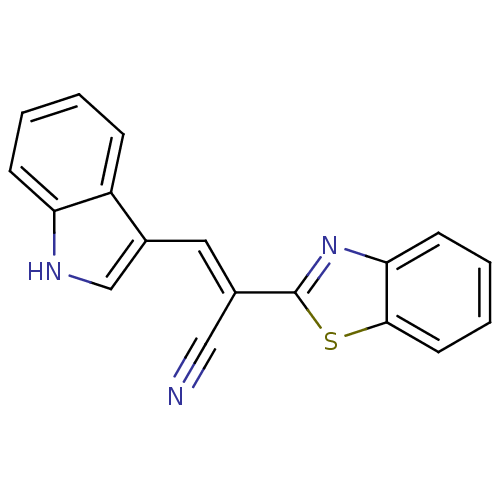
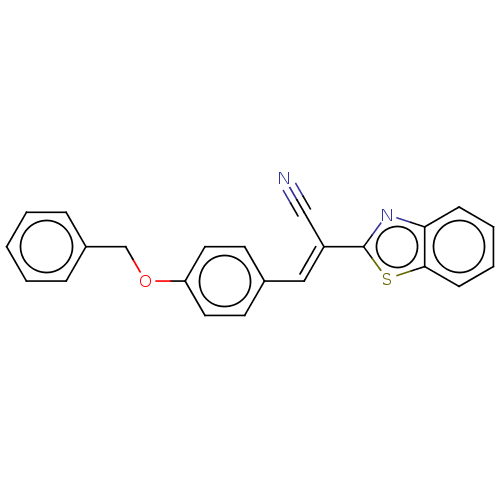
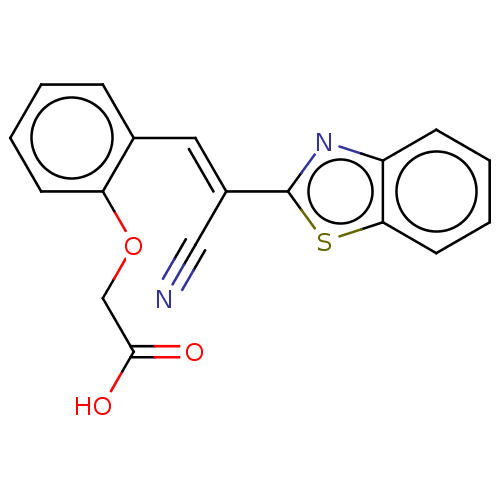
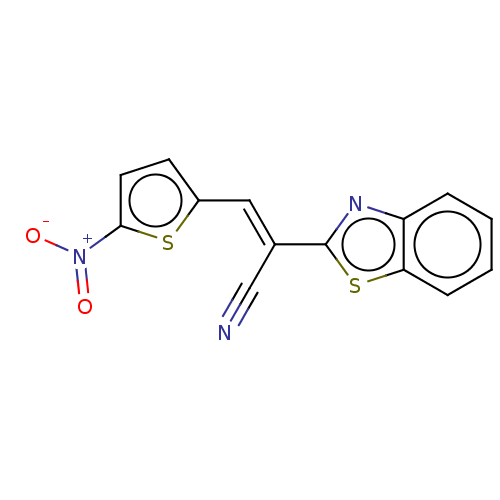
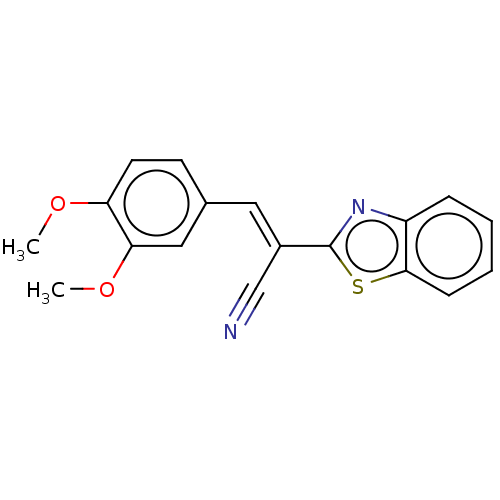
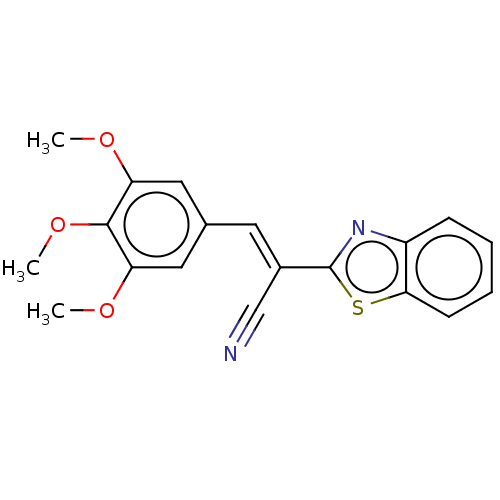
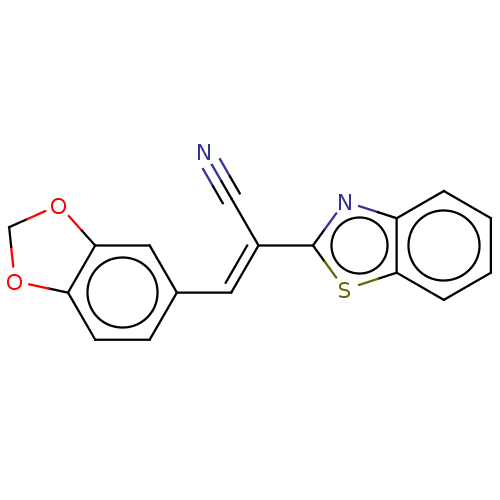
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 1.04E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine and alpha-ketoglutaric acid as substrate by spectrophotometric analysisMore data for this Ligand-Target Pair
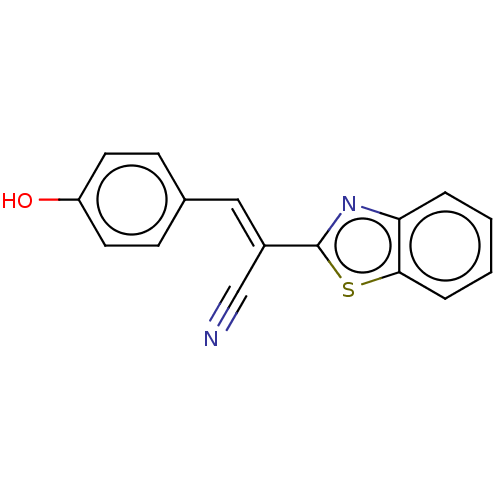
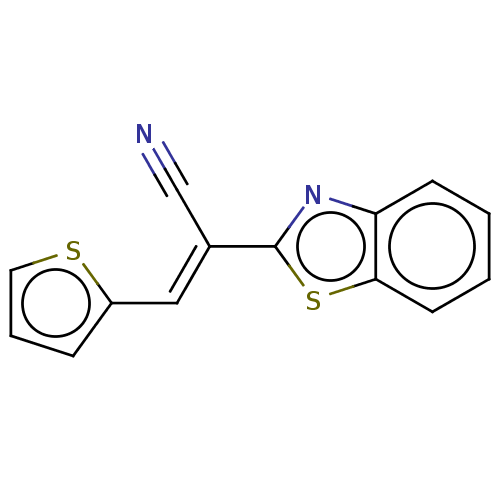
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 1.15E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
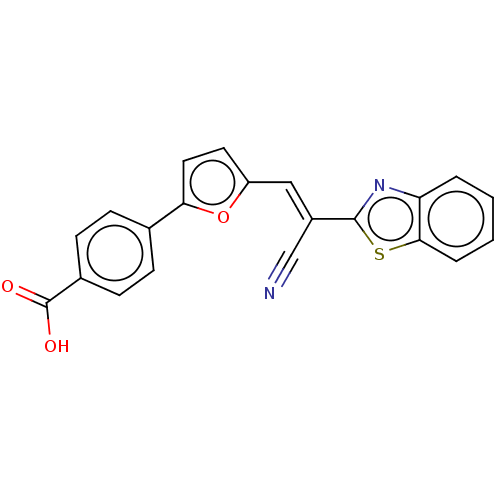
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 2.62E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
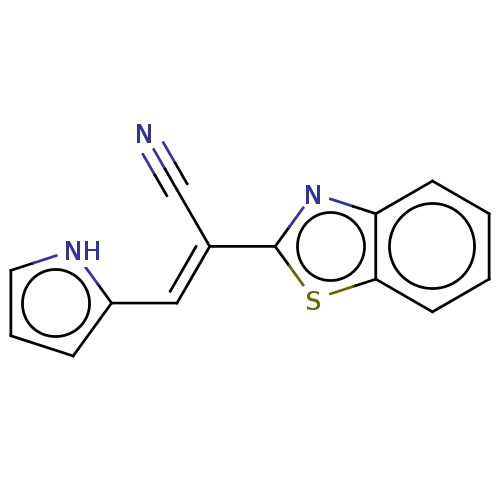
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 3.08E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 3.48E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine and alpha-ketoglutaric acid as substrate by spectrophotometric analysisMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 3.74E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 4.11E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 5.73E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 6.72E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 6.83E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine and alpha-ketoglutaric acid as substrate by spectrophotometric analysisMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 7.83E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 1.04E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 1.41E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 1.62E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 1.71E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 1.96E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 2.32E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 3.79E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 4.79E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 5.38E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 5.48E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 6.14E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 6.49E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 6.60E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair
TargetProbable L-lysine-epsilon aminotransferase(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
Institute of Technology & Science-Pilani
Curated by ChEMBL
Institute of Technology & Science-Pilani
Curated by ChEMBL
Affinity DataIC50: 9.26E+4nMAssay Description:Inhibition of Mycobacterium tuberculosis LAT using L-lysine as substrate by spectroscopic methodMore data for this Ligand-Target Pair