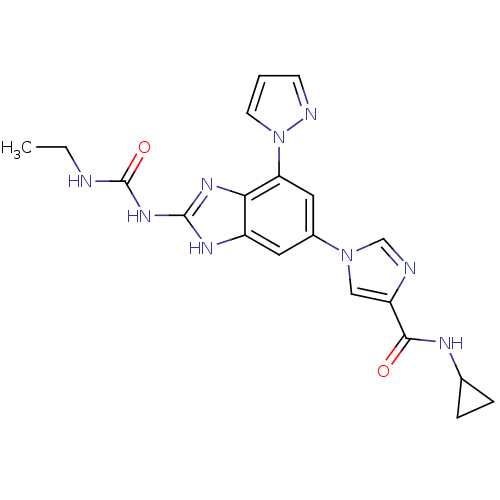
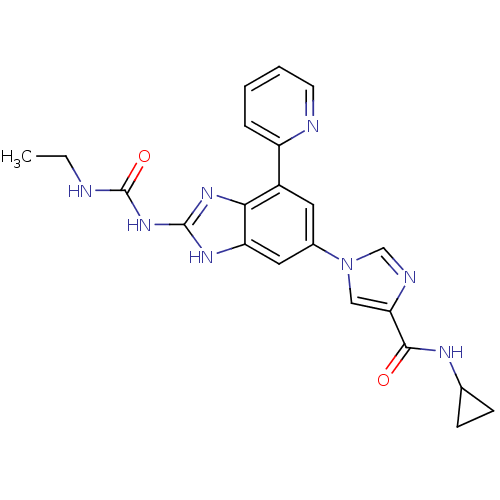
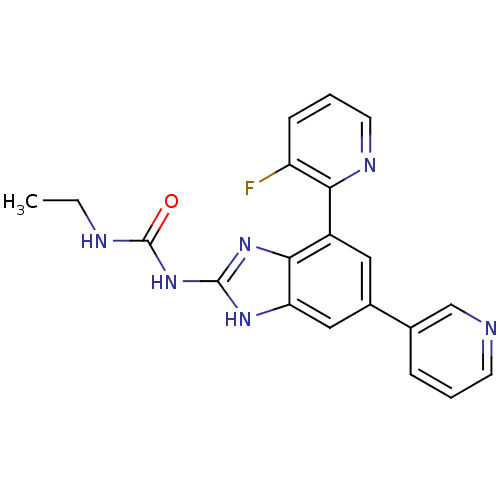
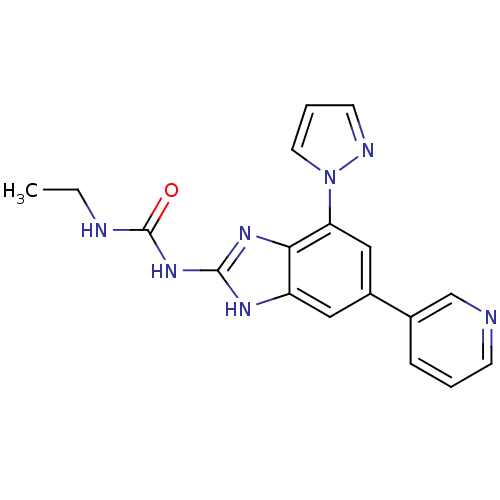
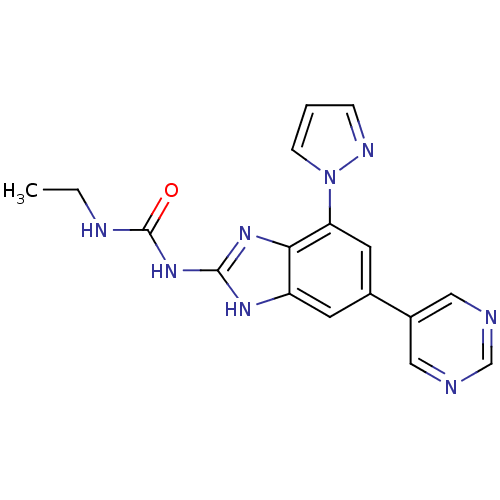
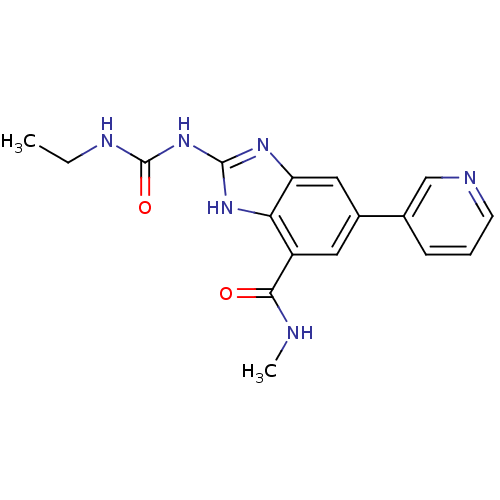
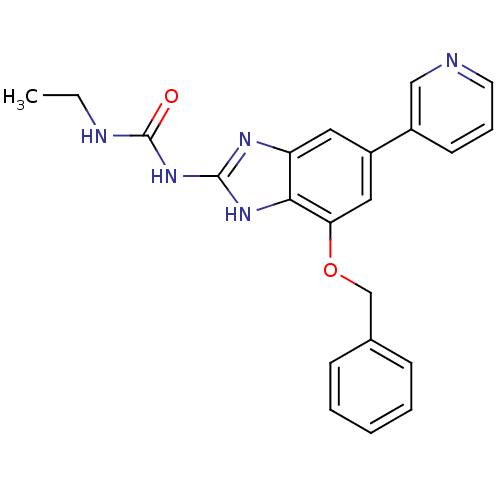
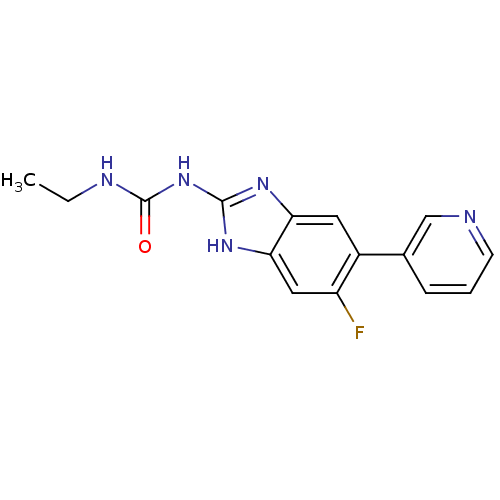
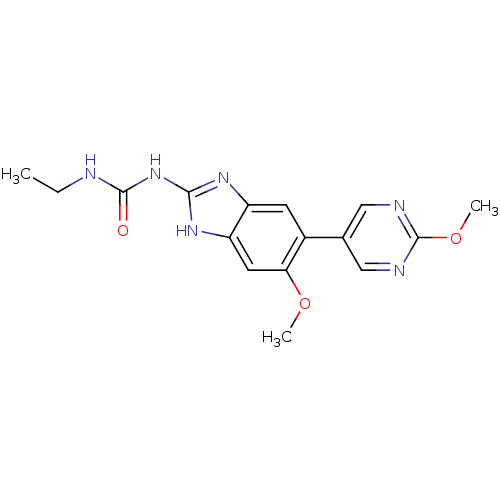
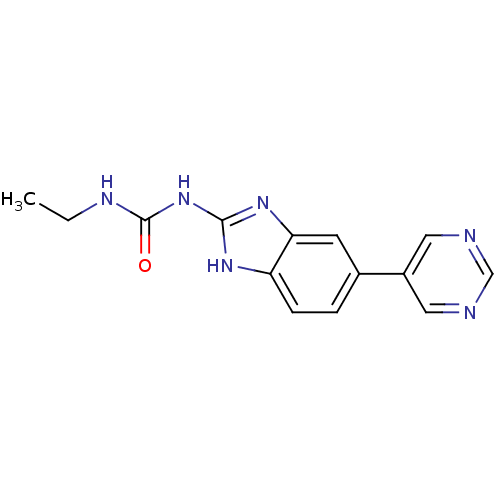
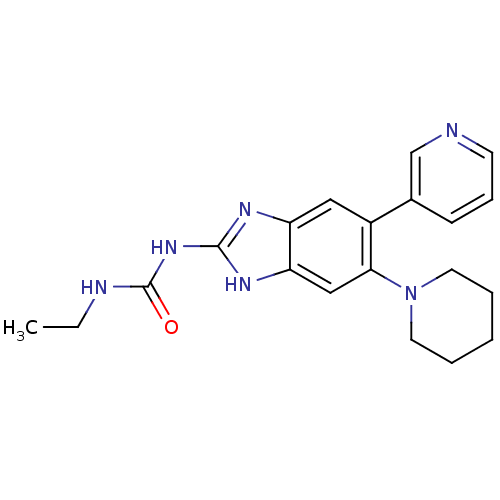
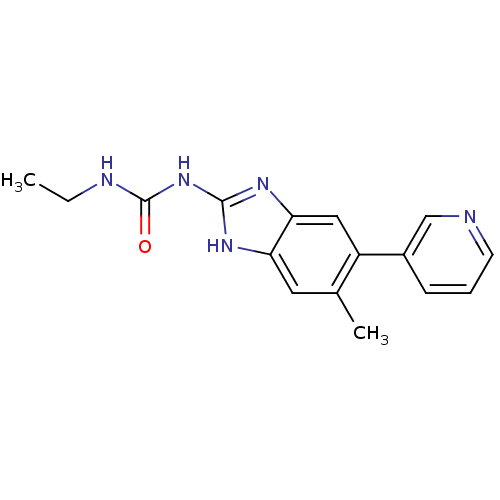
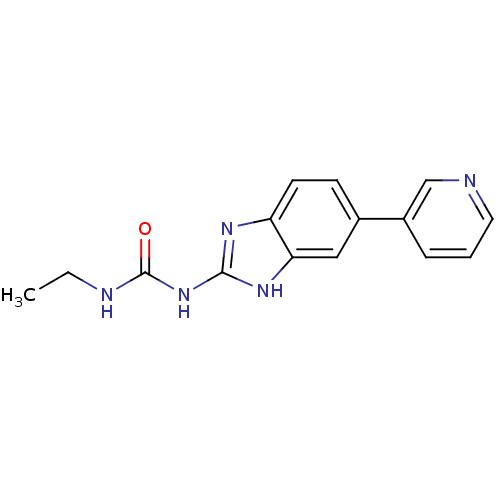
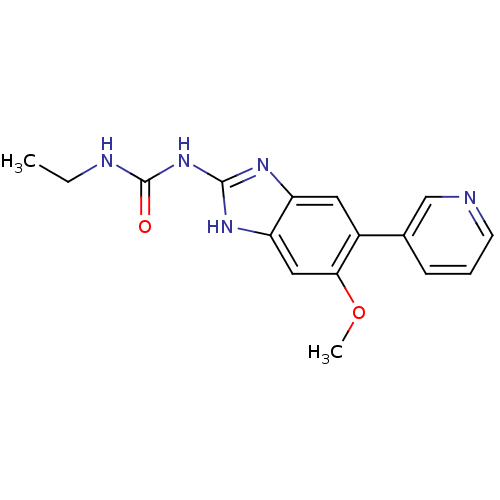
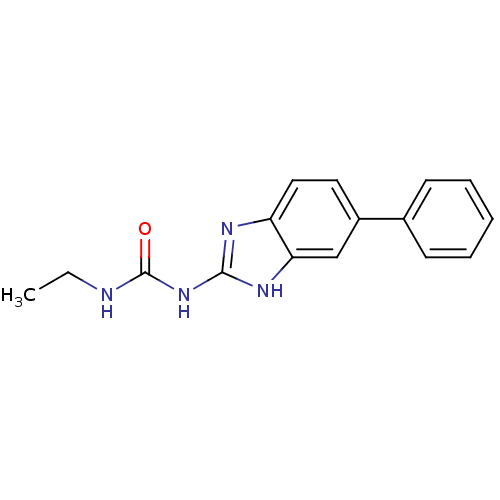
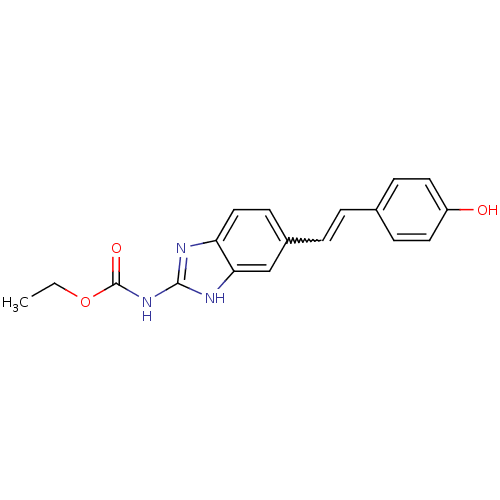
Affinity DataKi: 7nM ΔG°: -11.3kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
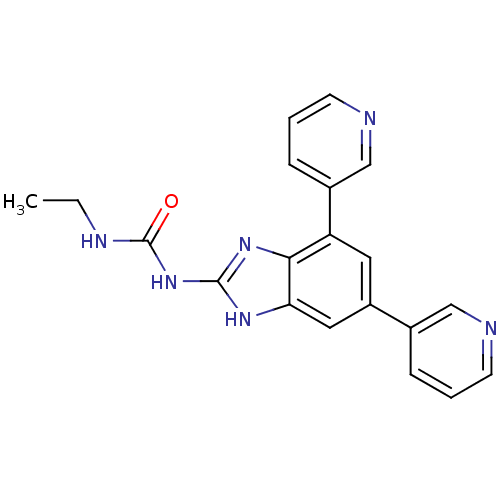
Affinity DataKi: 8nM ΔG°: -11.2kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
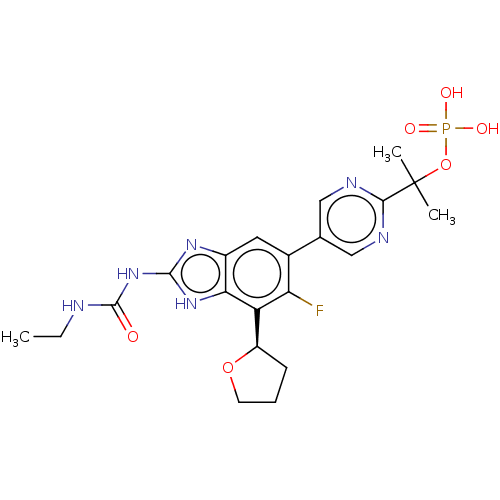
Affinity DataKi: 9nMAssay Description:Inhibition of Staphylococcus aureus DNA gyrase using pBR322 plasmid DNA as substrate by coupled enzyme reaction assayMore data for this Ligand-Target Pair
Affinity DataKi: 9nMpH: 7.6Assay Description:The ATP hydrolysis activity of S. aureus DNA gyrase is measured by coupling the production of ADP through pyruvate kinase/lactate dehydrogenase to th...More data for this Ligand-Target Pair
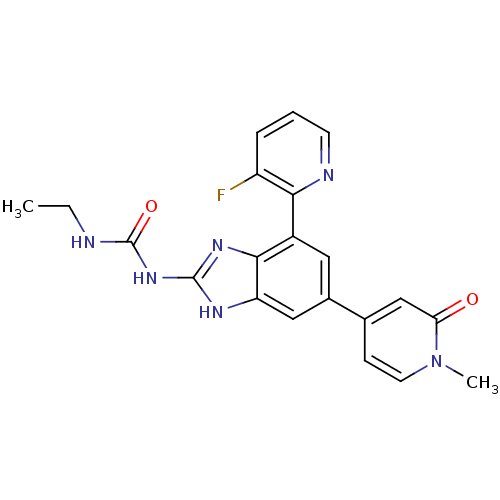
Affinity DataKi: 10nM ΔG°: -11.1kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nM ΔG°: -11.1kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 10nM ΔG°: -11.1kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 12nM ΔG°: -11.0kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 14nM ΔG°: -10.9kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 15nM ΔG°: -10.8kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 16nM ΔG°: -10.8kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 17nM ΔG°: -10.8kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: <17.7nMAssay Description:Inhibition of Staphylococcus aureus DNA gyraseMore data for this Ligand-Target Pair
Affinity DataKi: <21nMAssay Description:Inhibition of Staphylococcus aureus DNA gyraseMore data for this Ligand-Target Pair
Affinity DataKi: 23nM ΔG°: -10.6kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 26nM ΔG°: -10.5kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 35nM ΔG°: -10.3kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 38nM ΔG°: -10.3kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 44nM ΔG°: -10.2kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 49nM ΔG°: -10.1kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 81nM ΔG°: -9.83kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 85nM ΔG°: -9.80kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 130nM ΔG°: -9.54kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 190nM ΔG°: -9.32kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 470nM ΔG°: -8.77kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 2.00E+3nM ΔG°: -7.90kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair

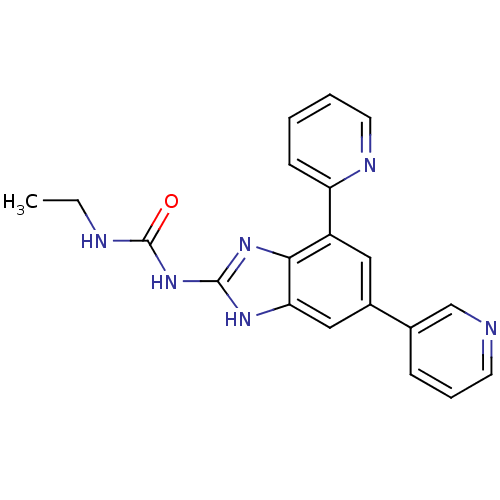
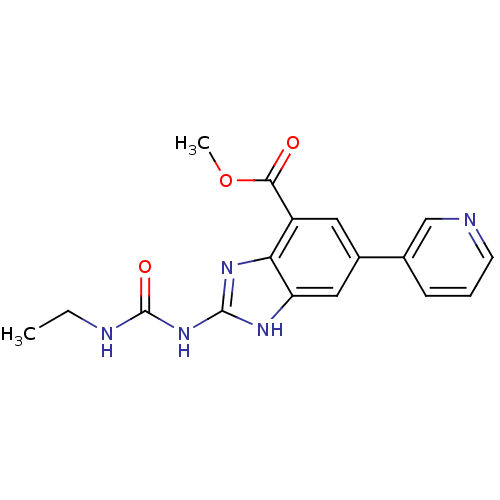
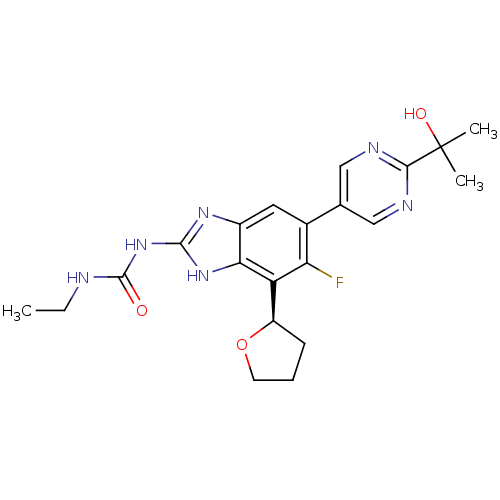
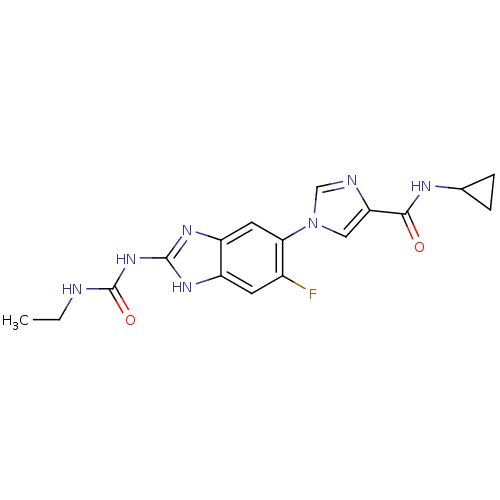
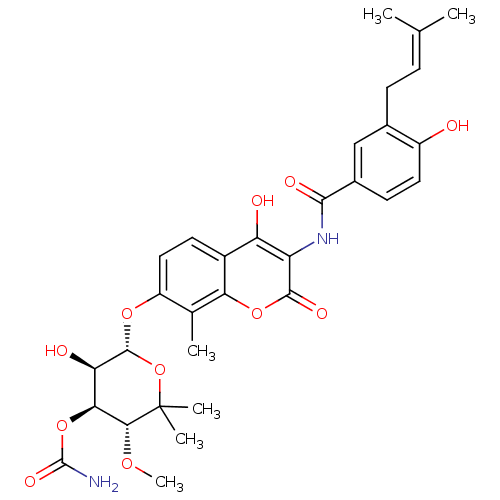
 3D Structure (crystal)
3D Structure (crystal)