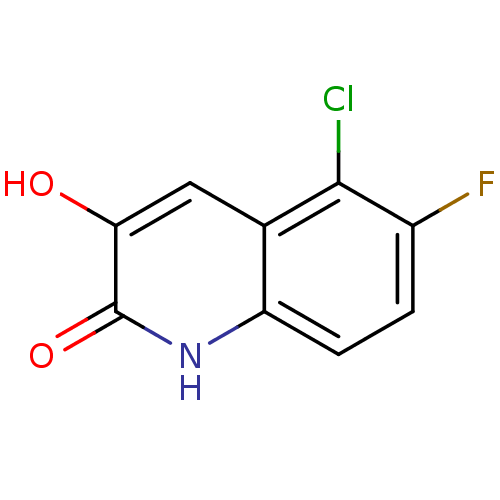
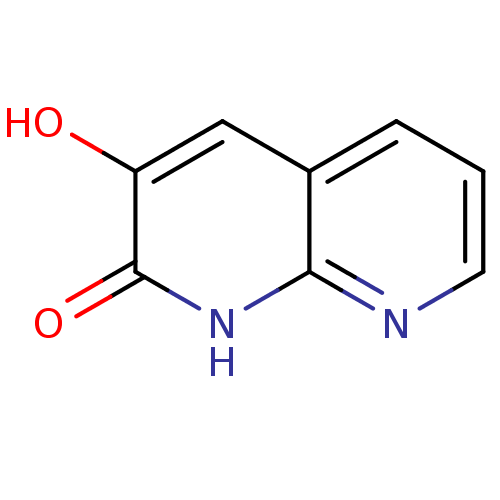
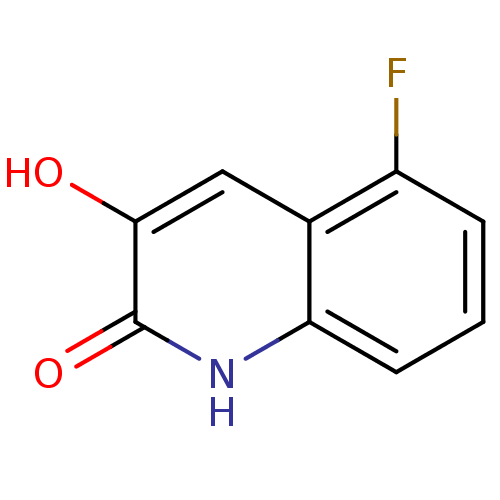
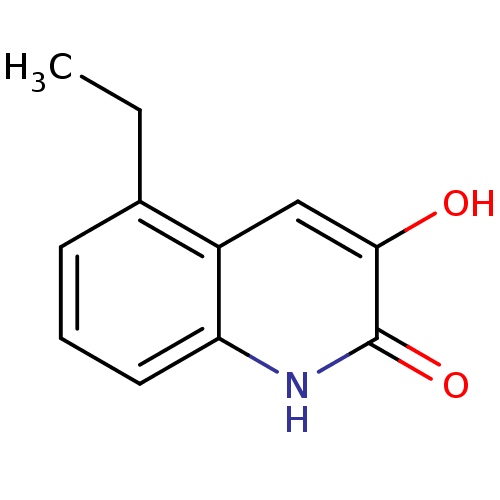
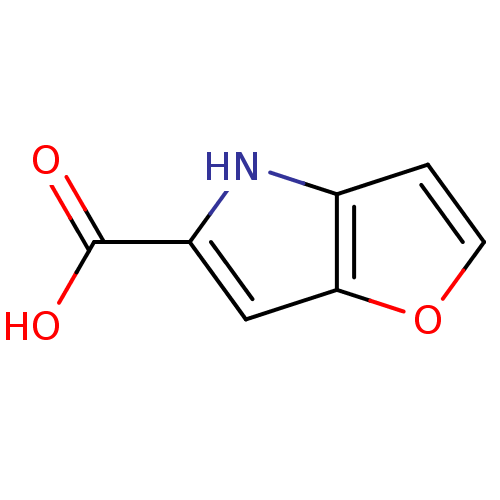
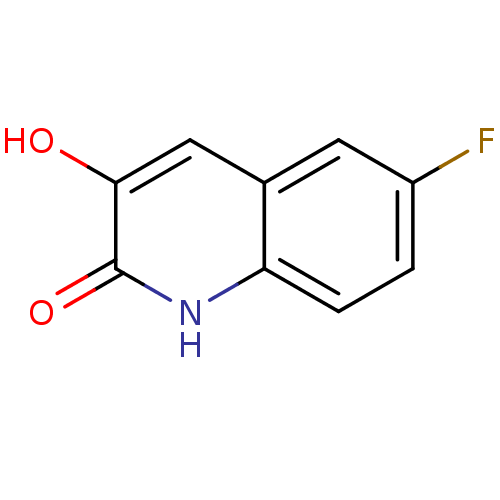
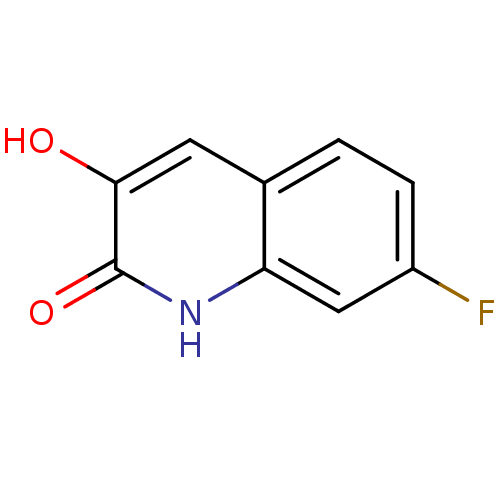
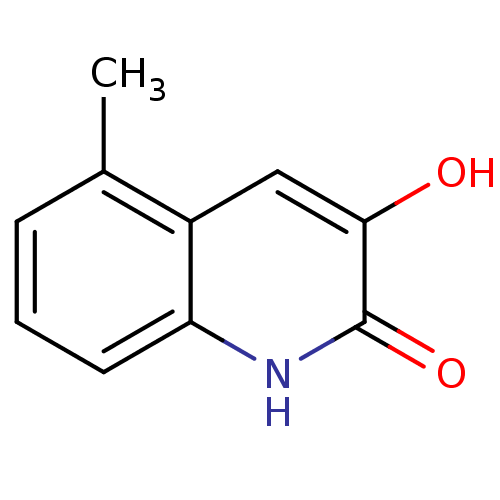
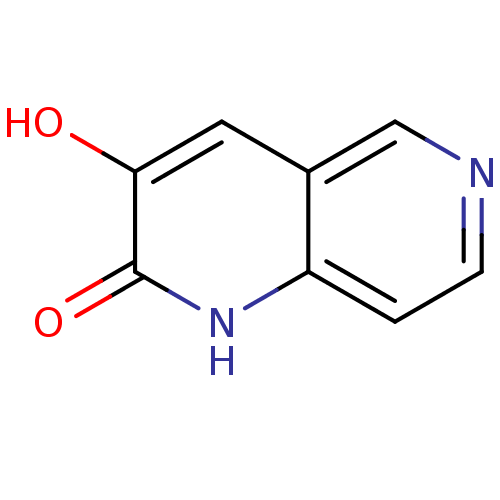
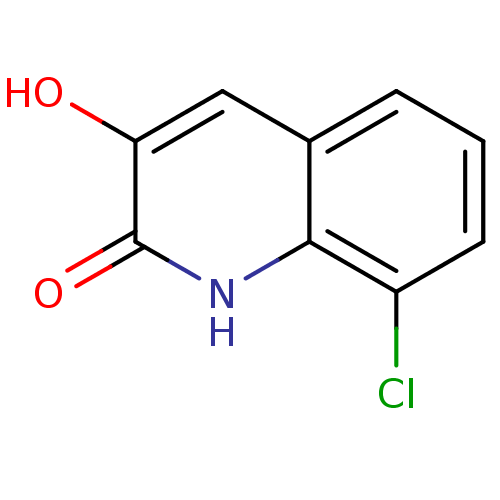
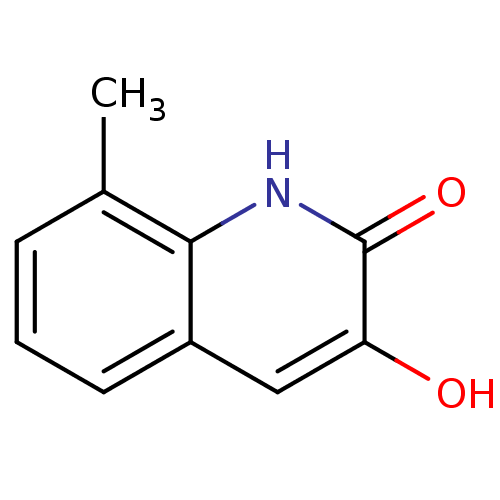
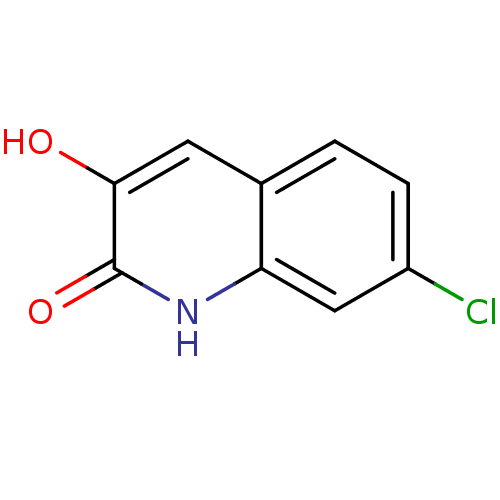
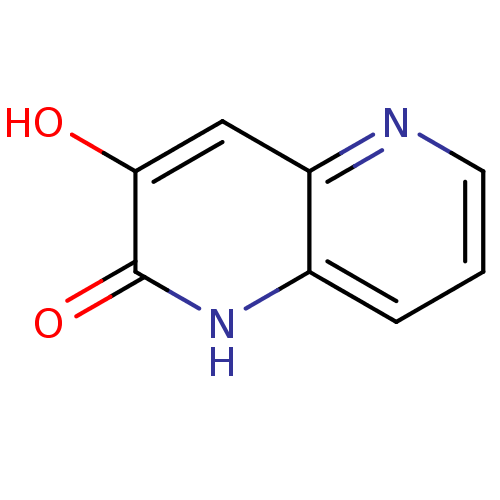
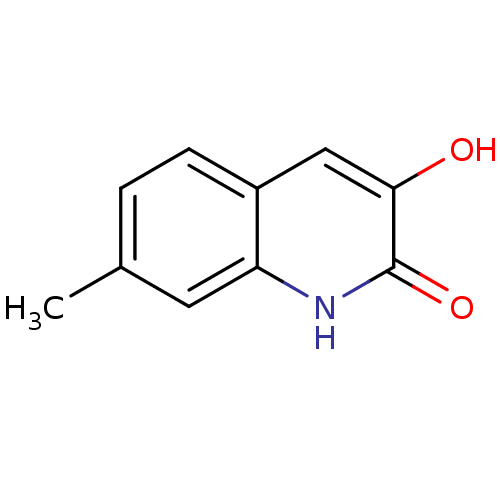
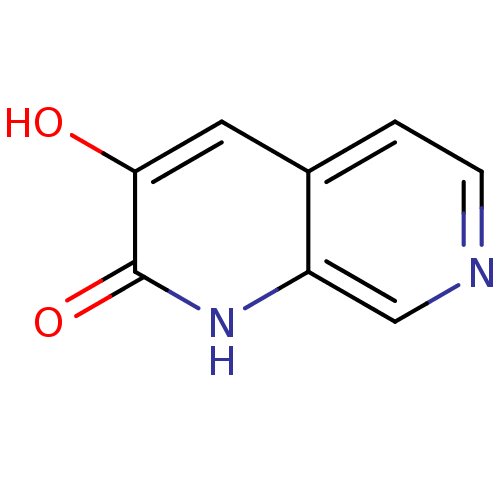
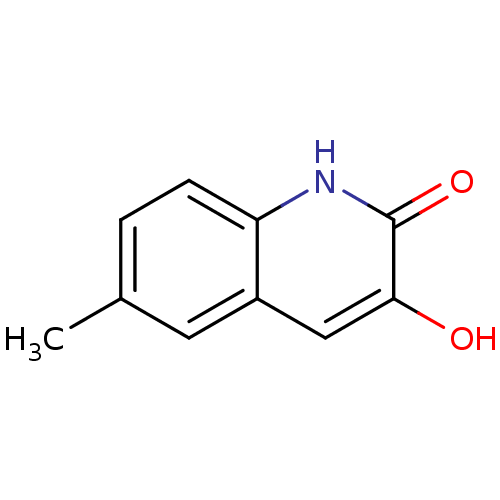
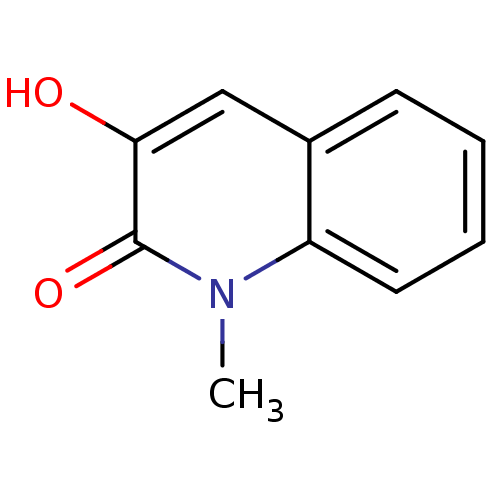
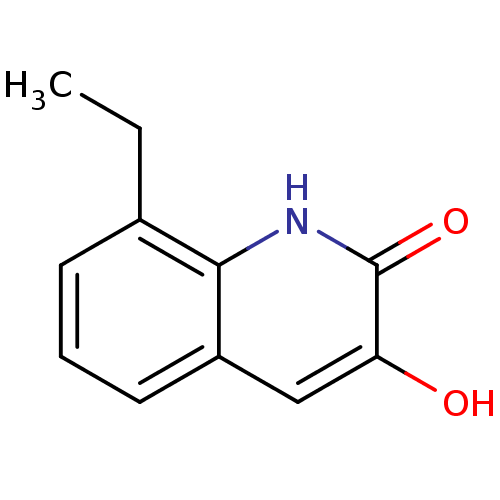
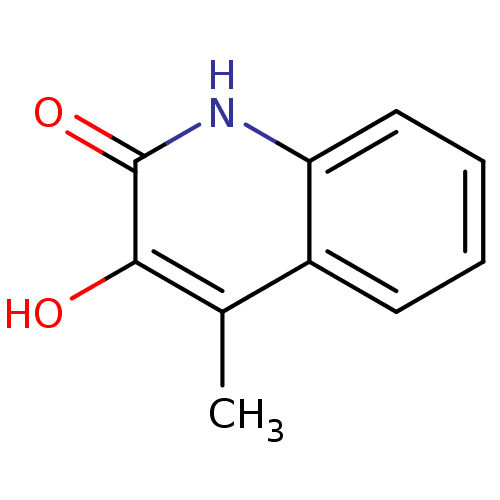
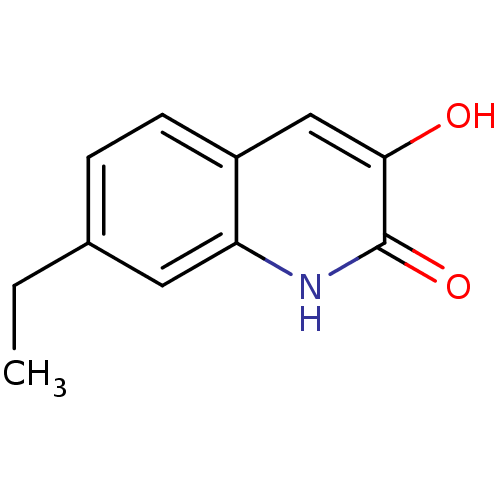
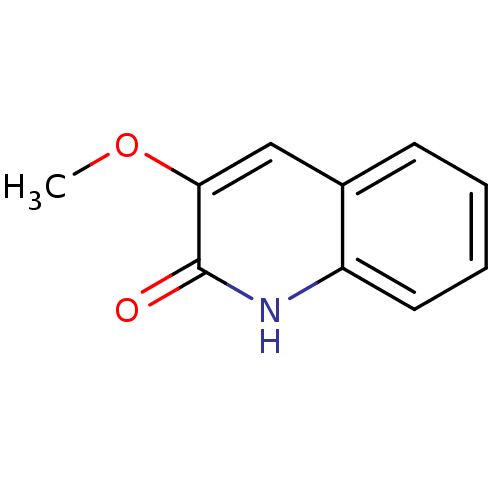
Affinity DataIC50: 3nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 3nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 5nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 8nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 8nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 8nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 9nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 9nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 10nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 16nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 32nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 33nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 38nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 40nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 50nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 100nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 128nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 155nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 180nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 196nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 197nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 215nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 308nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 424nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 784nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 855nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 1.45E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 2.75E+3nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 3.08E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 3.26E+3nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 3.67E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 4.62E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 7.64E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 8.47E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 8.72E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 8.88E+3nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 1.44E+4nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 1.45E+4nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 1.73E+4nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 1.95E+4nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 2.15E+4nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 2.24E+4nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 2.26E+4nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 2.62E+4nMpH: 8.5 T: 2°CAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-serine was linked to ox...More data for this Ligand-Target Pair
Affinity DataIC50: 3.16E+4nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair
Affinity DataIC50: 3.23E+4nMAssay Description:Inhibitory effect of compounds was determined in a cell free fluorescence assay. The H2O2 generated from the degradation of D-aspartic acid was linke...More data for this Ligand-Target Pair

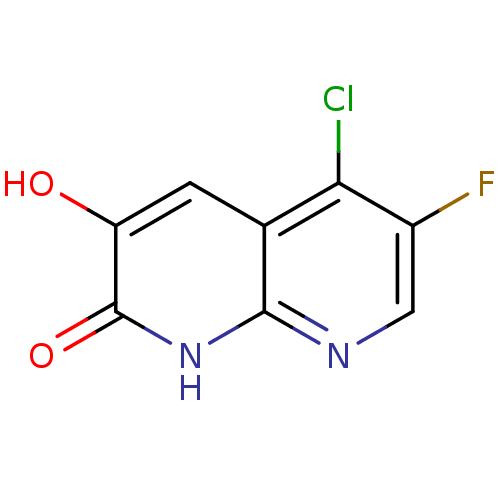
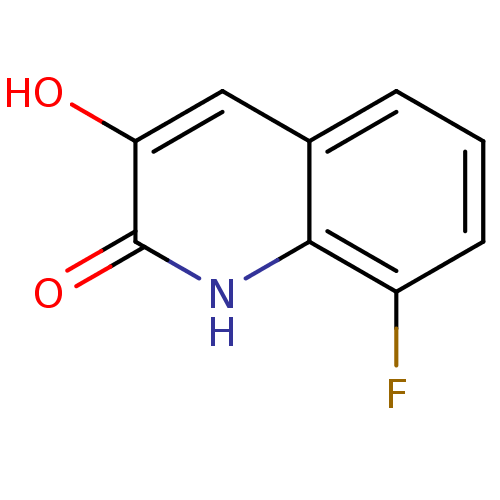
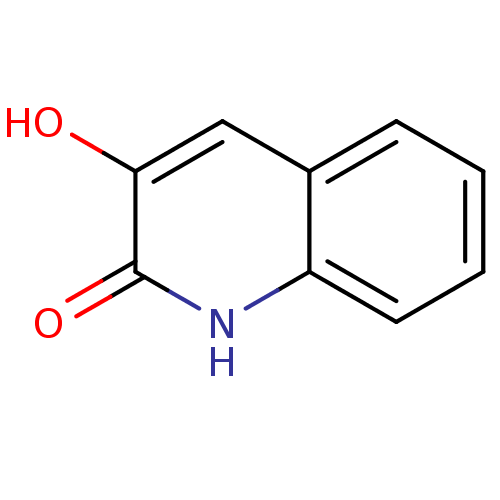
 3D Structure (crystal)
3D Structure (crystal)